
Anaerospora hongkongensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Negativicutes; Selenomonadales; Sporomusaceae; Anaerospora
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
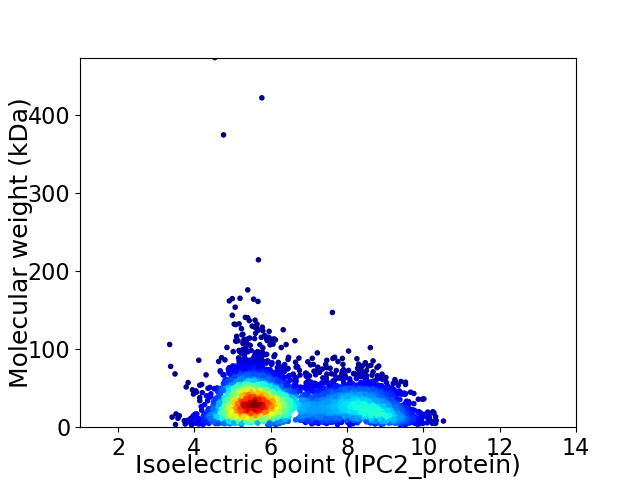
Virtual 2D-PAGE plot for 4497 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R1PXY2|A0A4R1PXY2_9FIRM Uncharacterized protein OS=Anaerospora hongkongensis OX=244830 GN=EV210_10564 PE=4 SV=1
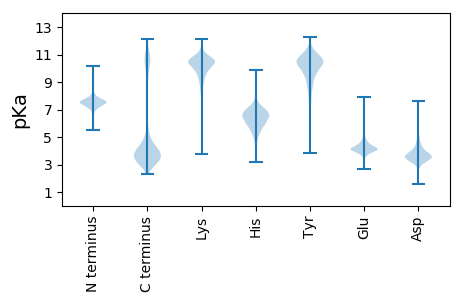
MM1 pKa = 7.59GFFDD5 pKa = 6.51DD6 pKa = 3.19ICNRR10 pKa = 11.84HH11 pKa = 4.58WCEE14 pKa = 4.5EE15 pKa = 4.34LEE17 pKa = 4.41CSCGINVVIFTADD30 pKa = 2.87GFAFFGLLDD39 pKa = 3.72RR40 pKa = 11.84VEE42 pKa = 4.72DD43 pKa = 4.68GIVILLPAAGEE54 pKa = 4.12NEE56 pKa = 4.31VFILTPGGDD65 pKa = 3.34VVTEE69 pKa = 3.81EE70 pKa = 3.99LSYY73 pKa = 11.06IEE75 pKa = 4.93LCTIVALSKK84 pKa = 11.04NVIGNPFEE92 pKa = 5.22EE93 pKa = 5.25DD94 pKa = 3.11DD95 pKa = 4.14CEE97 pKa = 4.92RR98 pKa = 11.84GGKK101 pKa = 8.98
MM1 pKa = 7.59GFFDD5 pKa = 6.51DD6 pKa = 3.19ICNRR10 pKa = 11.84HH11 pKa = 4.58WCEE14 pKa = 4.5EE15 pKa = 4.34LEE17 pKa = 4.41CSCGINVVIFTADD30 pKa = 2.87GFAFFGLLDD39 pKa = 3.72RR40 pKa = 11.84VEE42 pKa = 4.72DD43 pKa = 4.68GIVILLPAAGEE54 pKa = 4.12NEE56 pKa = 4.31VFILTPGGDD65 pKa = 3.34VVTEE69 pKa = 3.81EE70 pKa = 3.99LSYY73 pKa = 11.06IEE75 pKa = 4.93LCTIVALSKK84 pKa = 11.04NVIGNPFEE92 pKa = 5.22EE93 pKa = 5.25DD94 pKa = 3.11DD95 pKa = 4.14CEE97 pKa = 4.92RR98 pKa = 11.84GGKK101 pKa = 8.98
Molecular weight: 11.07 kDa
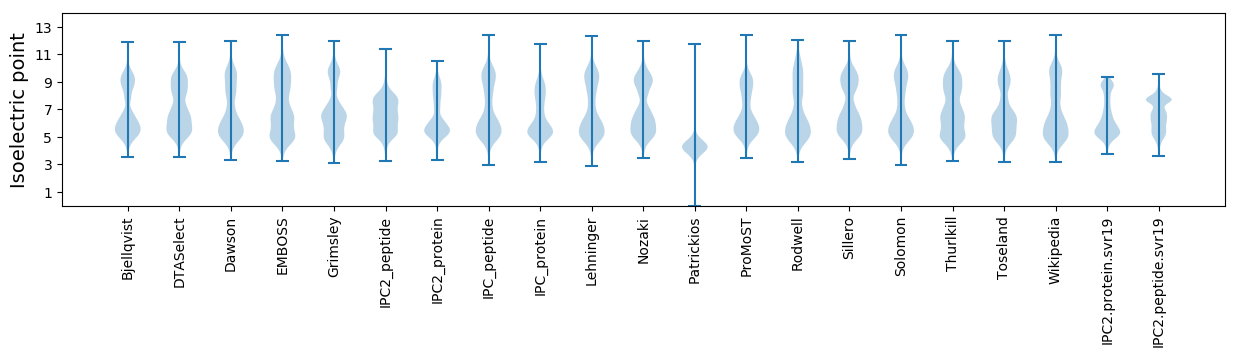
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R1Q3W3|A0A4R1Q3W3_9FIRM Nitrogen fixation NifU-like protein OS=Anaerospora hongkongensis OX=244830 GN=EV210_103191 PE=4 SV=1
MM1 pKa = 6.97KK2 pKa = 9.85QKK4 pKa = 11.44SMMALRR10 pKa = 11.84TIACGWLAFLCSFVDD25 pKa = 3.63RR26 pKa = 11.84LSWPPIIPMAAVEE39 pKa = 4.3LRR41 pKa = 11.84LTAAEE46 pKa = 4.31AGGFMSAFFFGYY58 pKa = 10.75LLTQLPGGMLADD70 pKa = 3.23RR71 pKa = 11.84WGTRR75 pKa = 11.84KK76 pKa = 9.79VLLVSLFVMGLFTMGLAVTNDD97 pKa = 3.58FGQGVILRR105 pKa = 11.84FLAGFGSGAVLAAAVKK121 pKa = 10.24GVYY124 pKa = 10.06DD125 pKa = 3.79HH126 pKa = 6.86VAAEE130 pKa = 4.08HH131 pKa = 6.58RR132 pKa = 11.84AMAMGFFMTSAPLGLLLANLLSPLIAAHH160 pKa = 5.57YY161 pKa = 8.94GWRR164 pKa = 11.84TSFLFAGSMTLAALGLTWLILPRR187 pKa = 11.84RR188 pKa = 11.84TGLRR192 pKa = 11.84IVPAAGKK199 pKa = 8.43TGIKK203 pKa = 10.22FEE205 pKa = 4.42LKK207 pKa = 10.28EE208 pKa = 4.14IITNRR213 pKa = 11.84EE214 pKa = 3.7LMLTAAAGFFAMWGTWGTLTWANAYY239 pKa = 7.4MHH241 pKa = 6.46QGLGLSLKK249 pKa = 10.42EE250 pKa = 4.3SGRR253 pKa = 11.84TMALFGIGALIGQPVVGWLSDD274 pKa = 3.23RR275 pKa = 11.84FRR277 pKa = 11.84QRR279 pKa = 11.84RR280 pKa = 11.84RR281 pKa = 11.84QASMVLLGVFAVLLWLFAKK300 pKa = 10.59NQDD303 pKa = 3.44AQYY306 pKa = 10.76LAVLATLLGAGAFMFGPVLNTFISEE331 pKa = 4.14LVAARR336 pKa = 11.84SVATAVGFCNGVWQIGSLISPVTAGLVLDD365 pKa = 3.75QTGSYY370 pKa = 10.54VYY372 pKa = 10.77AFSILAAGPLVAVGLLAFVRR392 pKa = 11.84SDD394 pKa = 2.94RR395 pKa = 11.84AFVEE399 pKa = 4.46RR400 pKa = 11.84PLTKK404 pKa = 10.46ARR406 pKa = 11.84IQAAGSLPGPVPKK419 pKa = 10.63
MM1 pKa = 6.97KK2 pKa = 9.85QKK4 pKa = 11.44SMMALRR10 pKa = 11.84TIACGWLAFLCSFVDD25 pKa = 3.63RR26 pKa = 11.84LSWPPIIPMAAVEE39 pKa = 4.3LRR41 pKa = 11.84LTAAEE46 pKa = 4.31AGGFMSAFFFGYY58 pKa = 10.75LLTQLPGGMLADD70 pKa = 3.23RR71 pKa = 11.84WGTRR75 pKa = 11.84KK76 pKa = 9.79VLLVSLFVMGLFTMGLAVTNDD97 pKa = 3.58FGQGVILRR105 pKa = 11.84FLAGFGSGAVLAAAVKK121 pKa = 10.24GVYY124 pKa = 10.06DD125 pKa = 3.79HH126 pKa = 6.86VAAEE130 pKa = 4.08HH131 pKa = 6.58RR132 pKa = 11.84AMAMGFFMTSAPLGLLLANLLSPLIAAHH160 pKa = 5.57YY161 pKa = 8.94GWRR164 pKa = 11.84TSFLFAGSMTLAALGLTWLILPRR187 pKa = 11.84RR188 pKa = 11.84TGLRR192 pKa = 11.84IVPAAGKK199 pKa = 8.43TGIKK203 pKa = 10.22FEE205 pKa = 4.42LKK207 pKa = 10.28EE208 pKa = 4.14IITNRR213 pKa = 11.84EE214 pKa = 3.7LMLTAAAGFFAMWGTWGTLTWANAYY239 pKa = 7.4MHH241 pKa = 6.46QGLGLSLKK249 pKa = 10.42EE250 pKa = 4.3SGRR253 pKa = 11.84TMALFGIGALIGQPVVGWLSDD274 pKa = 3.23RR275 pKa = 11.84FRR277 pKa = 11.84QRR279 pKa = 11.84RR280 pKa = 11.84RR281 pKa = 11.84QASMVLLGVFAVLLWLFAKK300 pKa = 10.59NQDD303 pKa = 3.44AQYY306 pKa = 10.76LAVLATLLGAGAFMFGPVLNTFISEE331 pKa = 4.14LVAARR336 pKa = 11.84SVATAVGFCNGVWQIGSLISPVTAGLVLDD365 pKa = 3.75QTGSYY370 pKa = 10.54VYY372 pKa = 10.77AFSILAAGPLVAVGLLAFVRR392 pKa = 11.84SDD394 pKa = 2.94RR395 pKa = 11.84AFVEE399 pKa = 4.46RR400 pKa = 11.84PLTKK404 pKa = 10.46ARR406 pKa = 11.84IQAAGSLPGPVPKK419 pKa = 10.63
Molecular weight: 44.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1371066 |
29 |
4707 |
304.9 |
33.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.646 ± 0.051 | 1.186 ± 0.018 |
4.76 ± 0.026 | 6.193 ± 0.036 |
3.813 ± 0.029 | 7.619 ± 0.045 |
1.877 ± 0.016 | 7.232 ± 0.036 |
5.505 ± 0.03 | 9.992 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.684 ± 0.021 | 3.826 ± 0.029 |
3.927 ± 0.023 | 4.074 ± 0.026 |
4.693 ± 0.031 | 5.605 ± 0.028 |
5.649 ± 0.041 | 7.499 ± 0.036 |
1.003 ± 0.014 | 3.218 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
