
Wenling tonguesole paramyxovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Cynoglossusvirus; Cynoglossus cynoglossusvirus
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
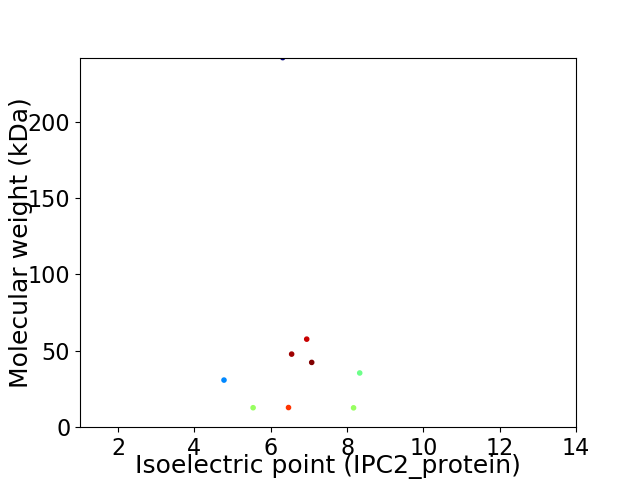
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1GMZ7|A0A2P1GMZ7_9MONO Uncharacterized protein OS=Wenling tonguesole paramyxovirus OX=2116454 PE=4 SV=1
MM1 pKa = 8.04DD2 pKa = 4.85WGDD5 pKa = 3.66EE6 pKa = 4.08QQTEE10 pKa = 4.22DD11 pKa = 3.55MFKK14 pKa = 10.06IGKK17 pKa = 8.38DD18 pKa = 3.35LKK20 pKa = 11.16VADD23 pKa = 3.97QMLSVNTHH31 pKa = 6.94DD32 pKa = 4.18SAEE35 pKa = 4.38DD36 pKa = 3.74YY37 pKa = 11.19ALSTSGVRR45 pKa = 11.84KK46 pKa = 9.87ILDD49 pKa = 3.4KK50 pKa = 11.42SGLTVTGSALLKK62 pKa = 10.15PGAKK66 pKa = 8.54STVISLFEE74 pKa = 4.31PLEE77 pKa = 4.13MAVSPEE83 pKa = 3.94PVQLVYY89 pKa = 10.65DD90 pKa = 4.03IPDD93 pKa = 3.29EE94 pKa = 4.72RR95 pKa = 11.84DD96 pKa = 3.42TIKK99 pKa = 8.12TTIEE103 pKa = 4.08DD104 pKa = 4.15PEE106 pKa = 4.74PNALLGEE113 pKa = 4.18IKK115 pKa = 10.29KK116 pKa = 9.77IQATQSQHH124 pKa = 4.69TSMLNEE130 pKa = 4.89IIQKK134 pKa = 10.07ISTQDD139 pKa = 3.49SILKK143 pKa = 9.78SISSISTQISILSTLLEE160 pKa = 4.98KK161 pKa = 10.09GTIMCANPALPDD173 pKa = 3.55QQSTRR178 pKa = 11.84LYY180 pKa = 10.8GATLEE185 pKa = 4.26EE186 pKa = 3.93NRR188 pKa = 11.84QEE190 pKa = 4.3KK191 pKa = 8.37MHH193 pKa = 6.49NLASRR198 pKa = 11.84APMFGALSEE207 pKa = 4.05ALKK210 pKa = 11.21NKK212 pKa = 8.87GKK214 pKa = 9.52PLIGYY219 pKa = 7.28GEE221 pKa = 4.36PSRR224 pKa = 11.84GSIMEE229 pKa = 5.25RR230 pKa = 11.84IPILLKK236 pKa = 10.61GLKK239 pKa = 8.51TSSRR243 pKa = 11.84NEE245 pKa = 3.73YY246 pKa = 8.89TSQYY250 pKa = 10.16VNRR253 pKa = 11.84DD254 pKa = 3.21DD255 pKa = 5.39SFDD258 pKa = 5.06PIQFSAEE265 pKa = 4.02LEE267 pKa = 4.2QVLSQEE273 pKa = 4.14QADD276 pKa = 3.75ISTEE280 pKa = 3.78
MM1 pKa = 8.04DD2 pKa = 4.85WGDD5 pKa = 3.66EE6 pKa = 4.08QQTEE10 pKa = 4.22DD11 pKa = 3.55MFKK14 pKa = 10.06IGKK17 pKa = 8.38DD18 pKa = 3.35LKK20 pKa = 11.16VADD23 pKa = 3.97QMLSVNTHH31 pKa = 6.94DD32 pKa = 4.18SAEE35 pKa = 4.38DD36 pKa = 3.74YY37 pKa = 11.19ALSTSGVRR45 pKa = 11.84KK46 pKa = 9.87ILDD49 pKa = 3.4KK50 pKa = 11.42SGLTVTGSALLKK62 pKa = 10.15PGAKK66 pKa = 8.54STVISLFEE74 pKa = 4.31PLEE77 pKa = 4.13MAVSPEE83 pKa = 3.94PVQLVYY89 pKa = 10.65DD90 pKa = 4.03IPDD93 pKa = 3.29EE94 pKa = 4.72RR95 pKa = 11.84DD96 pKa = 3.42TIKK99 pKa = 8.12TTIEE103 pKa = 4.08DD104 pKa = 4.15PEE106 pKa = 4.74PNALLGEE113 pKa = 4.18IKK115 pKa = 10.29KK116 pKa = 9.77IQATQSQHH124 pKa = 4.69TSMLNEE130 pKa = 4.89IIQKK134 pKa = 10.07ISTQDD139 pKa = 3.49SILKK143 pKa = 9.78SISSISTQISILSTLLEE160 pKa = 4.98KK161 pKa = 10.09GTIMCANPALPDD173 pKa = 3.55QQSTRR178 pKa = 11.84LYY180 pKa = 10.8GATLEE185 pKa = 4.26EE186 pKa = 3.93NRR188 pKa = 11.84QEE190 pKa = 4.3KK191 pKa = 8.37MHH193 pKa = 6.49NLASRR198 pKa = 11.84APMFGALSEE207 pKa = 4.05ALKK210 pKa = 11.21NKK212 pKa = 8.87GKK214 pKa = 9.52PLIGYY219 pKa = 7.28GEE221 pKa = 4.36PSRR224 pKa = 11.84GSIMEE229 pKa = 5.25RR230 pKa = 11.84IPILLKK236 pKa = 10.61GLKK239 pKa = 8.51TSSRR243 pKa = 11.84NEE245 pKa = 3.73YY246 pKa = 8.89TSQYY250 pKa = 10.16VNRR253 pKa = 11.84DD254 pKa = 3.21DD255 pKa = 5.39SFDD258 pKa = 5.06PIQFSAEE265 pKa = 4.02LEE267 pKa = 4.2QVLSQEE273 pKa = 4.14QADD276 pKa = 3.75ISTEE280 pKa = 3.78
Molecular weight: 30.85 kDa
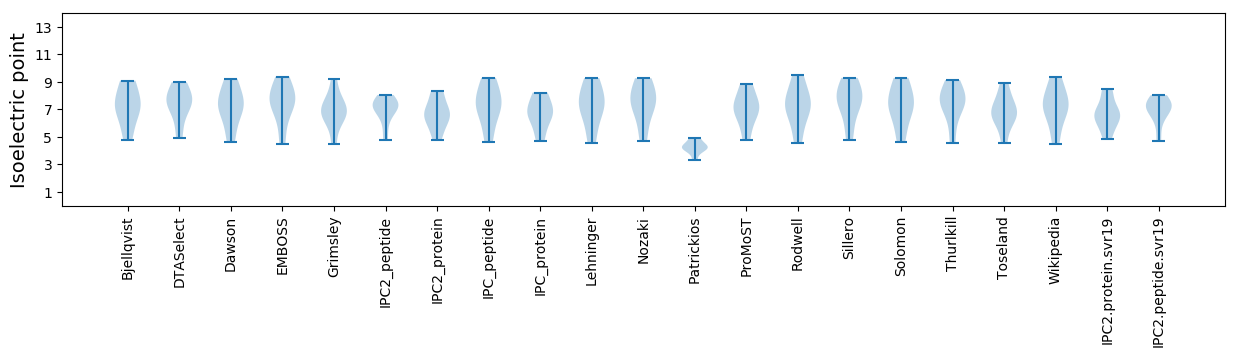
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1GMZ6|A0A2P1GMZ6_9MONO Uncharacterized protein OS=Wenling tonguesole paramyxovirus OX=2116454 PE=4 SV=1
MM1 pKa = 7.3EE2 pKa = 5.06VDD4 pKa = 4.84LGRR7 pKa = 11.84LPYY10 pKa = 10.25DD11 pKa = 3.2HH12 pKa = 7.74PGNDD16 pKa = 3.46RR17 pKa = 11.84DD18 pKa = 4.0KK19 pKa = 10.43LTGRR23 pKa = 11.84TGQLVLSPYY32 pKa = 10.84PNMTNDD38 pKa = 2.82RR39 pKa = 11.84VAIFVIGFVYY49 pKa = 10.24TGDD52 pKa = 3.21KK53 pKa = 10.32PAFNEE58 pKa = 4.33GEE60 pKa = 4.2KK61 pKa = 10.25LAKK64 pKa = 10.19ARR66 pKa = 11.84FPLGVHH72 pKa = 5.78NRR74 pKa = 11.84SDD76 pKa = 4.8DD77 pKa = 3.63FDD79 pKa = 5.96SYY81 pKa = 11.87TKK83 pKa = 10.65GLKK86 pKa = 10.32NSVFTVTRR94 pKa = 11.84VMRR97 pKa = 11.84KK98 pKa = 8.39VEE100 pKa = 4.14YY101 pKa = 10.12VALFPEE107 pKa = 4.56YY108 pKa = 9.84LDD110 pKa = 4.32PGIAPLYY117 pKa = 9.26PRR119 pKa = 11.84GKK121 pKa = 8.6RR122 pKa = 11.84TLMYY126 pKa = 10.14DD127 pKa = 2.9ARR129 pKa = 11.84KK130 pKa = 9.05FFPDD134 pKa = 2.59IHH136 pKa = 7.71HH137 pKa = 7.01FEE139 pKa = 4.16MNAPLKK145 pKa = 10.57CKK147 pKa = 9.21VVCVIYY153 pKa = 9.6TKK155 pKa = 10.35IPSAVKK161 pKa = 9.06IWKK164 pKa = 7.38PTFFSKK170 pKa = 10.99VEE172 pKa = 4.04SLPLVAMKK180 pKa = 10.33LVVEE184 pKa = 4.37IQLGEE189 pKa = 4.04IDD191 pKa = 3.71QGTAFTSHH199 pKa = 5.72IQIPLGWKK207 pKa = 9.19KK208 pKa = 11.02GKK210 pKa = 10.59GDD212 pKa = 5.03LEE214 pKa = 6.35DD215 pKa = 4.5DD216 pKa = 4.62LKK218 pKa = 11.48DD219 pKa = 3.5ALITIRR225 pKa = 11.84DD226 pKa = 3.74AAPKK230 pKa = 9.13VNLGAVHH237 pKa = 7.26GISLHH242 pKa = 6.26VMLTGQVSKK251 pKa = 10.84SIKK254 pKa = 10.23AYY256 pKa = 10.85LLGKK260 pKa = 9.38SAIVMSVADD269 pKa = 3.98TCPSVNFEE277 pKa = 4.76LCTGSISIKK286 pKa = 9.24SVKK289 pKa = 10.26ACATPTIPATAVLINSATEE308 pKa = 3.73DD309 pKa = 3.94LEE311 pKa = 4.23NHH313 pKa = 6.71AEE315 pKa = 4.12KK316 pKa = 10.44QKK318 pKa = 11.16KK319 pKa = 8.44NRR321 pKa = 3.7
MM1 pKa = 7.3EE2 pKa = 5.06VDD4 pKa = 4.84LGRR7 pKa = 11.84LPYY10 pKa = 10.25DD11 pKa = 3.2HH12 pKa = 7.74PGNDD16 pKa = 3.46RR17 pKa = 11.84DD18 pKa = 4.0KK19 pKa = 10.43LTGRR23 pKa = 11.84TGQLVLSPYY32 pKa = 10.84PNMTNDD38 pKa = 2.82RR39 pKa = 11.84VAIFVIGFVYY49 pKa = 10.24TGDD52 pKa = 3.21KK53 pKa = 10.32PAFNEE58 pKa = 4.33GEE60 pKa = 4.2KK61 pKa = 10.25LAKK64 pKa = 10.19ARR66 pKa = 11.84FPLGVHH72 pKa = 5.78NRR74 pKa = 11.84SDD76 pKa = 4.8DD77 pKa = 3.63FDD79 pKa = 5.96SYY81 pKa = 11.87TKK83 pKa = 10.65GLKK86 pKa = 10.32NSVFTVTRR94 pKa = 11.84VMRR97 pKa = 11.84KK98 pKa = 8.39VEE100 pKa = 4.14YY101 pKa = 10.12VALFPEE107 pKa = 4.56YY108 pKa = 9.84LDD110 pKa = 4.32PGIAPLYY117 pKa = 9.26PRR119 pKa = 11.84GKK121 pKa = 8.6RR122 pKa = 11.84TLMYY126 pKa = 10.14DD127 pKa = 2.9ARR129 pKa = 11.84KK130 pKa = 9.05FFPDD134 pKa = 2.59IHH136 pKa = 7.71HH137 pKa = 7.01FEE139 pKa = 4.16MNAPLKK145 pKa = 10.57CKK147 pKa = 9.21VVCVIYY153 pKa = 9.6TKK155 pKa = 10.35IPSAVKK161 pKa = 9.06IWKK164 pKa = 7.38PTFFSKK170 pKa = 10.99VEE172 pKa = 4.04SLPLVAMKK180 pKa = 10.33LVVEE184 pKa = 4.37IQLGEE189 pKa = 4.04IDD191 pKa = 3.71QGTAFTSHH199 pKa = 5.72IQIPLGWKK207 pKa = 9.19KK208 pKa = 11.02GKK210 pKa = 10.59GDD212 pKa = 5.03LEE214 pKa = 6.35DD215 pKa = 4.5DD216 pKa = 4.62LKK218 pKa = 11.48DD219 pKa = 3.5ALITIRR225 pKa = 11.84DD226 pKa = 3.74AAPKK230 pKa = 9.13VNLGAVHH237 pKa = 7.26GISLHH242 pKa = 6.26VMLTGQVSKK251 pKa = 10.84SIKK254 pKa = 10.23AYY256 pKa = 10.85LLGKK260 pKa = 9.38SAIVMSVADD269 pKa = 3.98TCPSVNFEE277 pKa = 4.76LCTGSISIKK286 pKa = 9.24SVKK289 pKa = 10.26ACATPTIPATAVLINSATEE308 pKa = 3.73DD309 pKa = 3.94LEE311 pKa = 4.23NHH313 pKa = 6.71AEE315 pKa = 4.12KK316 pKa = 10.44QKK318 pKa = 11.16KK319 pKa = 8.44NRR321 pKa = 3.7
Molecular weight: 35.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4415 |
110 |
2125 |
490.6 |
54.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.614 ± 0.812 | 1.789 ± 0.261 |
5.776 ± 0.346 | 4.96 ± 0.676 |
3.42 ± 0.344 | 5.685 ± 0.401 |
2.559 ± 0.357 | 7.746 ± 0.532 |
5.232 ± 0.576 | 10.51 ± 0.61 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.899 ± 0.356 | 4.643 ± 0.348 |
4.168 ± 0.309 | 3.692 ± 0.327 |
5.3 ± 0.572 | 7.407 ± 0.554 |
7.61 ± 0.59 | 5.798 ± 0.552 |
1.427 ± 0.382 | 2.763 ± 0.253 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
