
Acidovorax sp. SLBN-42
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae; Acidovorax; unclassified Acidovorax
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
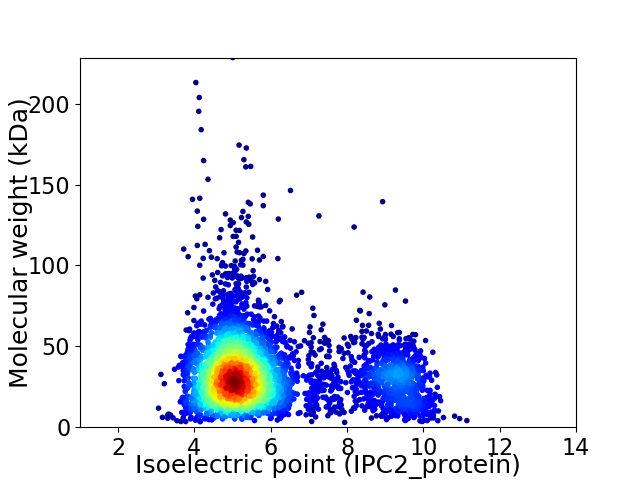
Virtual 2D-PAGE plot for 4383 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A542J5D8|A0A542J5D8_9BURK Biotin-dependent carboxylase-like uncharacterized protein OS=Acidovorax sp. SLBN-42 OX=2768435 GN=FBY15_1682 PE=4 SV=1
MM1 pKa = 7.02SAKK4 pKa = 9.19TGRR7 pKa = 11.84VRR9 pKa = 11.84SIGRR13 pKa = 11.84FSALVGATAVVAALGGCAQGGPVGFDD39 pKa = 3.3GVQSATIQLEE49 pKa = 4.08AVGTFVSPEE58 pKa = 3.75EE59 pKa = 4.31GGYY62 pKa = 9.39EE63 pKa = 3.71AAGRR67 pKa = 11.84GSGFIITSDD76 pKa = 3.03GLAITNNHH84 pKa = 5.19VVTGAGTLKK93 pKa = 10.3AWRR96 pKa = 11.84GGDD99 pKa = 3.39TTNEE103 pKa = 3.57LSAKK107 pKa = 9.88VLGSSEE113 pKa = 4.32CLDD116 pKa = 3.65LAVIQLEE123 pKa = 4.35SGTYY127 pKa = 9.97PFLAWRR133 pKa = 11.84DD134 pKa = 3.58DD135 pKa = 4.64DD136 pKa = 5.39IEE138 pKa = 4.32TALDD142 pKa = 3.55VYY144 pKa = 11.28AAGFPLGDD152 pKa = 3.52PTFTEE157 pKa = 4.33TKK159 pKa = 10.46GIVSKK164 pKa = 11.33ANTNGEE170 pKa = 4.42TPWASIDD177 pKa = 3.73GVIEE181 pKa = 3.46HH182 pKa = 7.56DD183 pKa = 3.26ARR185 pKa = 11.84IRR187 pKa = 11.84GGNSGGPLVDD197 pKa = 3.02TDD199 pKa = 3.41GRR201 pKa = 11.84LVGVNYY207 pKa = 10.54AGNDD211 pKa = 3.53EE212 pKa = 5.19LDD214 pKa = 3.56YY215 pKa = 11.32NYY217 pKa = 10.57AIHH220 pKa = 7.35RR221 pKa = 11.84DD222 pKa = 3.45AVLEE226 pKa = 4.3VVSDD230 pKa = 3.89LVDD233 pKa = 3.99GKK235 pKa = 10.72DD236 pKa = 3.61VLSLGINGEE245 pKa = 4.07ALTDD249 pKa = 3.99DD250 pKa = 4.35EE251 pKa = 4.84GTGLGVWVNSVEE263 pKa = 4.35SGSAADD269 pKa = 3.75EE270 pKa = 4.73AGVEE274 pKa = 4.21PGDD277 pKa = 3.78LLTRR281 pKa = 11.84MEE283 pKa = 4.63GVSLAANGTMTEE295 pKa = 4.17YY296 pKa = 11.06CDD298 pKa = 3.38VLRR301 pKa = 11.84THH303 pKa = 6.51GQDD306 pKa = 2.94DD307 pKa = 3.99TLAVEE312 pKa = 5.1LYY314 pKa = 7.99RR315 pKa = 11.84TSEE318 pKa = 3.77DD319 pKa = 2.87AYY321 pKa = 11.5YY322 pKa = 10.51RR323 pKa = 11.84GQFNGDD329 pKa = 4.16PITAVPVVGGGGTNEE344 pKa = 3.81PSGDD348 pKa = 3.9YY349 pKa = 9.62VTVTDD354 pKa = 4.68DD355 pKa = 3.44SQTITVEE362 pKa = 4.55VPTAWSQVDD371 pKa = 4.25GAAVPPWEE379 pKa = 4.75SVTAAPDD386 pKa = 3.34LAGYY390 pKa = 7.3RR391 pKa = 11.84TTWATPGVTVYY402 pKa = 10.86ASQTAGVTPDD412 pKa = 2.98EE413 pKa = 4.52AANLVTEE420 pKa = 4.52GLAAQGCTSAGRR432 pKa = 11.84EE433 pKa = 4.01PYY435 pKa = 10.66DD436 pKa = 3.93DD437 pKa = 3.84GLYY440 pKa = 10.64AGVWEE445 pKa = 4.76IFTGCGGQATYY456 pKa = 11.19VVIGANDD463 pKa = 3.52YY464 pKa = 11.4DD465 pKa = 4.02GTHH468 pKa = 6.91TIVFTGQAITDD479 pKa = 3.92ADD481 pKa = 4.84LDD483 pKa = 4.36ALDD486 pKa = 3.72QAIGTFYY493 pKa = 11.05AAYY496 pKa = 10.21
MM1 pKa = 7.02SAKK4 pKa = 9.19TGRR7 pKa = 11.84VRR9 pKa = 11.84SIGRR13 pKa = 11.84FSALVGATAVVAALGGCAQGGPVGFDD39 pKa = 3.3GVQSATIQLEE49 pKa = 4.08AVGTFVSPEE58 pKa = 3.75EE59 pKa = 4.31GGYY62 pKa = 9.39EE63 pKa = 3.71AAGRR67 pKa = 11.84GSGFIITSDD76 pKa = 3.03GLAITNNHH84 pKa = 5.19VVTGAGTLKK93 pKa = 10.3AWRR96 pKa = 11.84GGDD99 pKa = 3.39TTNEE103 pKa = 3.57LSAKK107 pKa = 9.88VLGSSEE113 pKa = 4.32CLDD116 pKa = 3.65LAVIQLEE123 pKa = 4.35SGTYY127 pKa = 9.97PFLAWRR133 pKa = 11.84DD134 pKa = 3.58DD135 pKa = 4.64DD136 pKa = 5.39IEE138 pKa = 4.32TALDD142 pKa = 3.55VYY144 pKa = 11.28AAGFPLGDD152 pKa = 3.52PTFTEE157 pKa = 4.33TKK159 pKa = 10.46GIVSKK164 pKa = 11.33ANTNGEE170 pKa = 4.42TPWASIDD177 pKa = 3.73GVIEE181 pKa = 3.46HH182 pKa = 7.56DD183 pKa = 3.26ARR185 pKa = 11.84IRR187 pKa = 11.84GGNSGGPLVDD197 pKa = 3.02TDD199 pKa = 3.41GRR201 pKa = 11.84LVGVNYY207 pKa = 10.54AGNDD211 pKa = 3.53EE212 pKa = 5.19LDD214 pKa = 3.56YY215 pKa = 11.32NYY217 pKa = 10.57AIHH220 pKa = 7.35RR221 pKa = 11.84DD222 pKa = 3.45AVLEE226 pKa = 4.3VVSDD230 pKa = 3.89LVDD233 pKa = 3.99GKK235 pKa = 10.72DD236 pKa = 3.61VLSLGINGEE245 pKa = 4.07ALTDD249 pKa = 3.99DD250 pKa = 4.35EE251 pKa = 4.84GTGLGVWVNSVEE263 pKa = 4.35SGSAADD269 pKa = 3.75EE270 pKa = 4.73AGVEE274 pKa = 4.21PGDD277 pKa = 3.78LLTRR281 pKa = 11.84MEE283 pKa = 4.63GVSLAANGTMTEE295 pKa = 4.17YY296 pKa = 11.06CDD298 pKa = 3.38VLRR301 pKa = 11.84THH303 pKa = 6.51GQDD306 pKa = 2.94DD307 pKa = 3.99TLAVEE312 pKa = 5.1LYY314 pKa = 7.99RR315 pKa = 11.84TSEE318 pKa = 3.77DD319 pKa = 2.87AYY321 pKa = 11.5YY322 pKa = 10.51RR323 pKa = 11.84GQFNGDD329 pKa = 4.16PITAVPVVGGGGTNEE344 pKa = 3.81PSGDD348 pKa = 3.9YY349 pKa = 9.62VTVTDD354 pKa = 4.68DD355 pKa = 3.44SQTITVEE362 pKa = 4.55VPTAWSQVDD371 pKa = 4.25GAAVPPWEE379 pKa = 4.75SVTAAPDD386 pKa = 3.34LAGYY390 pKa = 7.3RR391 pKa = 11.84TTWATPGVTVYY402 pKa = 10.86ASQTAGVTPDD412 pKa = 2.98EE413 pKa = 4.52AANLVTEE420 pKa = 4.52GLAAQGCTSAGRR432 pKa = 11.84EE433 pKa = 4.01PYY435 pKa = 10.66DD436 pKa = 3.93DD437 pKa = 3.84GLYY440 pKa = 10.64AGVWEE445 pKa = 4.76IFTGCGGQATYY456 pKa = 11.19VVIGANDD463 pKa = 3.52YY464 pKa = 11.4DD465 pKa = 4.02GTHH468 pKa = 6.91TIVFTGQAITDD479 pKa = 3.92ADD481 pKa = 4.84LDD483 pKa = 4.36ALDD486 pKa = 3.72QAIGTFYY493 pKa = 11.05AAYY496 pKa = 10.21
Molecular weight: 51.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A542J1A6|A0A542J1A6_9BURK ABC-2 type transport system ATP-binding protein OS=Acidovorax sp. SLBN-42 OX=2768435 GN=FBY15_0173 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1418679 |
29 |
2104 |
323.7 |
34.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.909 ± 0.048 | 0.483 ± 0.008 |
6.2 ± 0.031 | 5.946 ± 0.03 |
3.163 ± 0.019 | 9.056 ± 0.032 |
2.055 ± 0.018 | 4.377 ± 0.028 |
1.67 ± 0.022 | 10.119 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.78 ± 0.014 | 1.839 ± 0.016 |
5.526 ± 0.027 | 2.532 ± 0.021 |
7.616 ± 0.045 | 5.331 ± 0.027 |
5.775 ± 0.027 | 9.105 ± 0.036 |
1.549 ± 0.016 | 1.969 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
