
Beihai mantis shrimp virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
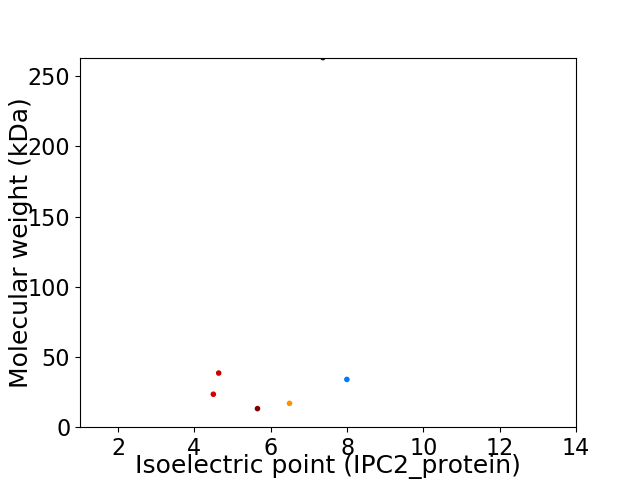
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJM5|A0A1L3KJM5_9VIRU Uncharacterized protein OS=Beihai mantis shrimp virus 1 OX=1922428 PE=4 SV=1
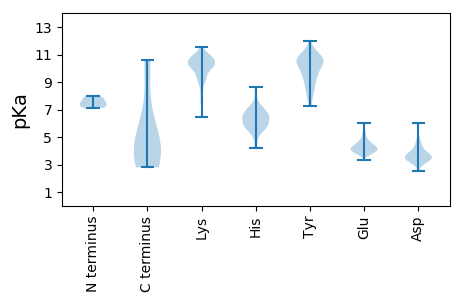
MM1 pKa = 7.81AFTEE5 pKa = 4.6LGWLTVKK12 pKa = 10.76KK13 pKa = 9.23LTQAPRR19 pKa = 11.84GLVVLLTTSLVVQQLMFLAVLNFNSLVMNQTAIIVEE55 pKa = 4.11NHH57 pKa = 5.98KK58 pKa = 11.23SLMEE62 pKa = 4.93DD63 pKa = 3.07LTSTQEE69 pKa = 4.15EE70 pKa = 4.42IASGITKK77 pKa = 10.3LDD79 pKa = 3.57GNVMEE84 pKa = 5.2FQLEE88 pKa = 4.48YY89 pKa = 9.78TNHH92 pKa = 5.98WPKK95 pKa = 10.85LDD97 pKa = 3.99PDD99 pKa = 4.21EE100 pKa = 4.82KK101 pKa = 10.81DD102 pKa = 2.91KK103 pKa = 11.45EE104 pKa = 4.5YY105 pKa = 11.22VSRR108 pKa = 11.84EE109 pKa = 3.86DD110 pKa = 3.4FDD112 pKa = 4.4EE113 pKa = 4.67FKK115 pKa = 11.14KK116 pKa = 10.88NYY118 pKa = 8.64AKK120 pKa = 10.44RR121 pKa = 11.84WLAEE125 pKa = 3.8PEE127 pKa = 4.23GEE129 pKa = 4.21YY130 pKa = 9.4VTSIKK135 pKa = 10.33FVEE138 pKa = 4.56KK139 pKa = 10.07LDD141 pKa = 3.83EE142 pKa = 4.77VIALLKK148 pKa = 10.68VPLVEE153 pKa = 4.27KK154 pKa = 10.75LNAVVEE160 pKa = 4.48TMEE163 pKa = 5.06FPACQTVSWDD173 pKa = 3.82GVLCEE178 pKa = 4.86NDD180 pKa = 3.31PFGIGPINCDD190 pKa = 2.92EE191 pKa = 4.18VVYY194 pKa = 10.84EE195 pKa = 4.39GTTCEE200 pKa = 4.01KK201 pKa = 10.51PVFIGII207 pKa = 4.02
MM1 pKa = 7.81AFTEE5 pKa = 4.6LGWLTVKK12 pKa = 10.76KK13 pKa = 9.23LTQAPRR19 pKa = 11.84GLVVLLTTSLVVQQLMFLAVLNFNSLVMNQTAIIVEE55 pKa = 4.11NHH57 pKa = 5.98KK58 pKa = 11.23SLMEE62 pKa = 4.93DD63 pKa = 3.07LTSTQEE69 pKa = 4.15EE70 pKa = 4.42IASGITKK77 pKa = 10.3LDD79 pKa = 3.57GNVMEE84 pKa = 5.2FQLEE88 pKa = 4.48YY89 pKa = 9.78TNHH92 pKa = 5.98WPKK95 pKa = 10.85LDD97 pKa = 3.99PDD99 pKa = 4.21EE100 pKa = 4.82KK101 pKa = 10.81DD102 pKa = 2.91KK103 pKa = 11.45EE104 pKa = 4.5YY105 pKa = 11.22VSRR108 pKa = 11.84EE109 pKa = 3.86DD110 pKa = 3.4FDD112 pKa = 4.4EE113 pKa = 4.67FKK115 pKa = 11.14KK116 pKa = 10.88NYY118 pKa = 8.64AKK120 pKa = 10.44RR121 pKa = 11.84WLAEE125 pKa = 3.8PEE127 pKa = 4.23GEE129 pKa = 4.21YY130 pKa = 9.4VTSIKK135 pKa = 10.33FVEE138 pKa = 4.56KK139 pKa = 10.07LDD141 pKa = 3.83EE142 pKa = 4.77VIALLKK148 pKa = 10.68VPLVEE153 pKa = 4.27KK154 pKa = 10.75LNAVVEE160 pKa = 4.48TMEE163 pKa = 5.06FPACQTVSWDD173 pKa = 3.82GVLCEE178 pKa = 4.86NDD180 pKa = 3.31PFGIGPINCDD190 pKa = 2.92EE191 pKa = 4.18VVYY194 pKa = 10.84EE195 pKa = 4.39GTTCEE200 pKa = 4.01KK201 pKa = 10.51PVFIGII207 pKa = 4.02
Molecular weight: 23.47 kDa
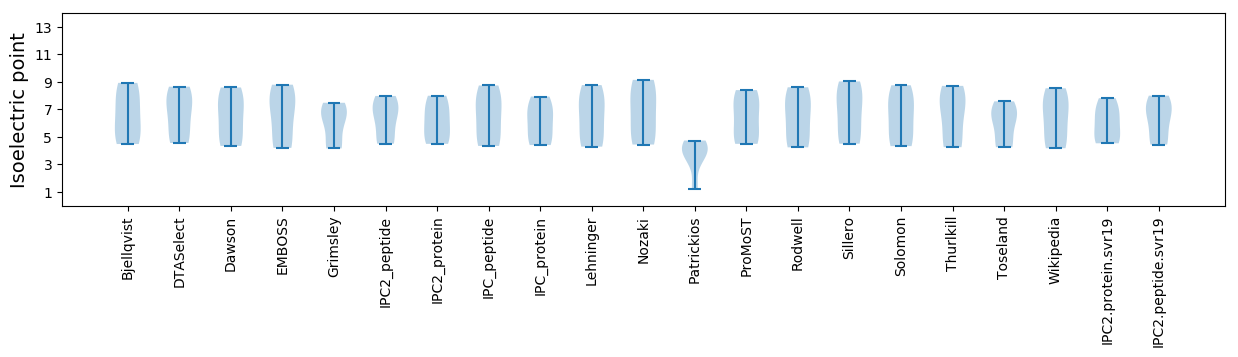
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJR8|A0A1L3KJR8_9VIRU Uncharacterized protein OS=Beihai mantis shrimp virus 1 OX=1922428 PE=4 SV=1
MM1 pKa = 7.54AFSFTWNKK9 pKa = 9.74LVQSKK14 pKa = 9.21MEE16 pKa = 4.12SSKK19 pKa = 11.42ALDD22 pKa = 3.33SDD24 pKa = 3.95FTAGKK29 pKa = 9.97RR30 pKa = 11.84EE31 pKa = 3.89VVLTSGALPASYY43 pKa = 9.76EE44 pKa = 4.16VKK46 pKa = 9.53VQKK49 pKa = 8.45TVRR52 pKa = 11.84ITGTDD57 pKa = 3.23NIGDD61 pKa = 3.91LKK63 pKa = 11.05LFLSLQPSIDD73 pKa = 3.7AACEE77 pKa = 3.98EE78 pKa = 3.87IAKK81 pKa = 10.14RR82 pKa = 11.84YY83 pKa = 8.11EE84 pKa = 4.1RR85 pKa = 11.84YY86 pKa = 10.58SIDD89 pKa = 4.03NANLWVQTASPLGTSSGGFQLAHH112 pKa = 6.63IPDD115 pKa = 4.46PEE117 pKa = 4.24NANLPAADD125 pKa = 4.17PANATKK131 pKa = 10.41AVRR134 pKa = 11.84QQGSSFIRR142 pKa = 11.84PRR144 pKa = 11.84DD145 pKa = 3.53SVAFPIPLQGQLYY158 pKa = 7.81TKK160 pKa = 10.4QAGSQRR166 pKa = 11.84WWSFGAIVGVVRR178 pKa = 11.84AVPDD182 pKa = 3.22ATDD185 pKa = 3.54YY186 pKa = 12.0VEE188 pKa = 4.78FTCTFTATFRR198 pKa = 11.84FMRR201 pKa = 11.84TAEE204 pKa = 3.72VDD206 pKa = 3.14NNQRR210 pKa = 11.84VAFRR214 pKa = 11.84TLISEE219 pKa = 4.19ATVEE223 pKa = 4.07EE224 pKa = 4.51GRR226 pKa = 11.84LRR228 pKa = 11.84LRR230 pKa = 11.84TTAPVPGRR238 pKa = 11.84RR239 pKa = 11.84GTVTLPRR246 pKa = 11.84PAAVEE251 pKa = 3.93VRR253 pKa = 11.84DD254 pKa = 3.8RR255 pKa = 11.84SARR258 pKa = 11.84GVIVSHH264 pKa = 6.39SRR266 pKa = 11.84TATFDD271 pKa = 4.09CIIEE275 pKa = 4.41GQCATSVRR283 pKa = 11.84DD284 pKa = 3.94DD285 pKa = 3.99FVALHH290 pKa = 6.2TPLTNSTCLFQLQRR304 pKa = 11.84TFYY307 pKa = 10.29CQQ309 pKa = 2.8
MM1 pKa = 7.54AFSFTWNKK9 pKa = 9.74LVQSKK14 pKa = 9.21MEE16 pKa = 4.12SSKK19 pKa = 11.42ALDD22 pKa = 3.33SDD24 pKa = 3.95FTAGKK29 pKa = 9.97RR30 pKa = 11.84EE31 pKa = 3.89VVLTSGALPASYY43 pKa = 9.76EE44 pKa = 4.16VKK46 pKa = 9.53VQKK49 pKa = 8.45TVRR52 pKa = 11.84ITGTDD57 pKa = 3.23NIGDD61 pKa = 3.91LKK63 pKa = 11.05LFLSLQPSIDD73 pKa = 3.7AACEE77 pKa = 3.98EE78 pKa = 3.87IAKK81 pKa = 10.14RR82 pKa = 11.84YY83 pKa = 8.11EE84 pKa = 4.1RR85 pKa = 11.84YY86 pKa = 10.58SIDD89 pKa = 4.03NANLWVQTASPLGTSSGGFQLAHH112 pKa = 6.63IPDD115 pKa = 4.46PEE117 pKa = 4.24NANLPAADD125 pKa = 4.17PANATKK131 pKa = 10.41AVRR134 pKa = 11.84QQGSSFIRR142 pKa = 11.84PRR144 pKa = 11.84DD145 pKa = 3.53SVAFPIPLQGQLYY158 pKa = 7.81TKK160 pKa = 10.4QAGSQRR166 pKa = 11.84WWSFGAIVGVVRR178 pKa = 11.84AVPDD182 pKa = 3.22ATDD185 pKa = 3.54YY186 pKa = 12.0VEE188 pKa = 4.78FTCTFTATFRR198 pKa = 11.84FMRR201 pKa = 11.84TAEE204 pKa = 3.72VDD206 pKa = 3.14NNQRR210 pKa = 11.84VAFRR214 pKa = 11.84TLISEE219 pKa = 4.19ATVEE223 pKa = 4.07EE224 pKa = 4.51GRR226 pKa = 11.84LRR228 pKa = 11.84LRR230 pKa = 11.84TTAPVPGRR238 pKa = 11.84RR239 pKa = 11.84GTVTLPRR246 pKa = 11.84PAAVEE251 pKa = 3.93VRR253 pKa = 11.84DD254 pKa = 3.8RR255 pKa = 11.84SARR258 pKa = 11.84GVIVSHH264 pKa = 6.39SRR266 pKa = 11.84TATFDD271 pKa = 4.09CIIEE275 pKa = 4.41GQCATSVRR283 pKa = 11.84DD284 pKa = 3.94DD285 pKa = 3.99FVALHH290 pKa = 6.2TPLTNSTCLFQLQRR304 pKa = 11.84TFYY307 pKa = 10.29CQQ309 pKa = 2.8
Molecular weight: 34.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3447 |
113 |
2318 |
574.5 |
64.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.253 ± 0.797 | 2.176 ± 0.414 |
5.86 ± 0.401 | 7.079 ± 0.813 |
4.352 ± 0.212 | 5.976 ± 0.4 |
2.263 ± 0.448 | 4.584 ± 0.105 |
6.934 ± 1.218 | 8.326 ± 0.464 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.553 ± 0.467 | 4.41 ± 0.58 |
4.062 ± 0.409 | 4.439 ± 0.384 |
5.628 ± 0.756 | 5.947 ± 0.417 |
5.454 ± 1.296 | 8.239 ± 0.44 |
1.247 ± 0.114 | 3.22 ± 0.582 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
