
Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Sordariomycetidae; Sordariales; Chaetomiaceae; Chaetomium; Chaetomium thermophilum; Chaetomium thermophilum var. thermophilum
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
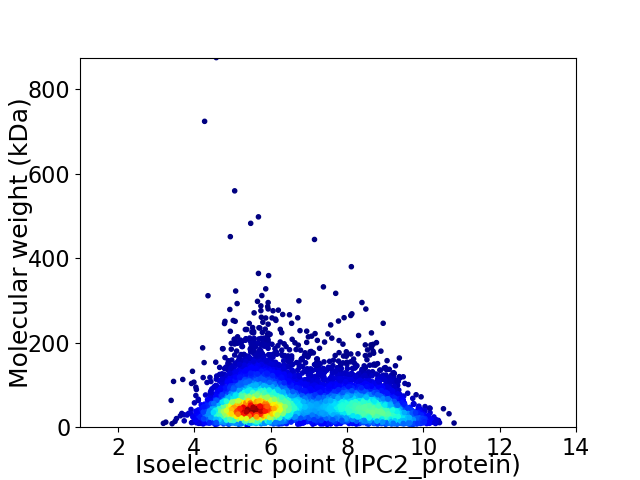
Virtual 2D-PAGE plot for 7185 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G0SBF1|G0SBF1_CHATD Beta-xylanase OS=Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) OX=759272 GN=CTHT_0050050 PE=3 SV=1
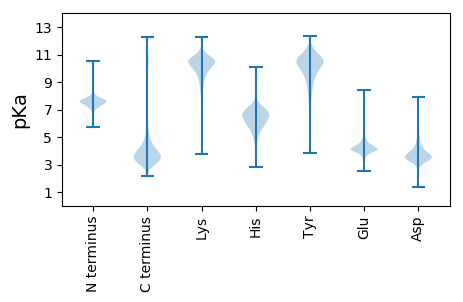
MM1 pKa = 7.86RR2 pKa = 11.84IWTSIACLSGMVSLATAAVVPRR24 pKa = 11.84VDD26 pKa = 4.75EE27 pKa = 4.43IALSSCTTSEE37 pKa = 3.91EE38 pKa = 4.52SVTVVTITVHH48 pKa = 5.83PQTATTITVTAGVSSPSSLSSALPFPFPGDD78 pKa = 3.27SSLGSEE84 pKa = 4.33STTCTDD90 pKa = 3.63EE91 pKa = 5.76DD92 pKa = 3.88SGSTDD97 pKa = 3.05VSIITVTPSLSWSFTFSTDD116 pKa = 3.26TTSSSADD123 pKa = 3.07AVTAGGPIQTVTVYY137 pKa = 10.48VPAPTPVTVTVGPPFSPSSASTVTSDD163 pKa = 3.48TEE165 pKa = 4.37CPVTITYY172 pKa = 7.78TLSPGPVFPSGFTGTLTLGGSKK194 pKa = 10.1PSSTPEE200 pKa = 3.52VSSAFDD206 pKa = 3.65PFSTFPEE213 pKa = 4.39SSTFGTPSSPILGPVTSSDD232 pKa = 3.4ADD234 pKa = 3.65SSDD237 pKa = 3.34VSVDD241 pKa = 3.17VVTYY245 pKa = 7.37TIPAQGTSSAQIGTVTVTSGAAQSTVSDD273 pKa = 3.83SVFTLTLSVFEE284 pKa = 4.93PSSPTYY290 pKa = 10.06NIPISSQPGTVTVTVTSAPDD310 pKa = 3.6DD311 pKa = 4.85DD312 pKa = 6.31GNDD315 pKa = 5.22DD316 pKa = 5.76DD317 pKa = 6.29DD318 pKa = 5.26DD319 pKa = 3.66WWTVWTNSWEE329 pKa = 4.39SISTITAIDD338 pKa = 3.72DD339 pKa = 3.7HH340 pKa = 8.2SSTVSSQDD348 pKa = 3.27ALSIWTSDD356 pKa = 2.91EE357 pKa = 3.72GATVVTVTEE366 pKa = 4.26IEE368 pKa = 4.35EE369 pKa = 4.2LTATPSSPGKK379 pKa = 9.23PWPTGYY385 pKa = 10.4DD386 pKa = 3.02SSGASVVVVTYY397 pKa = 7.17TIPGGSSTYY406 pKa = 9.56TVTYY410 pKa = 7.59TIPPLSTPSEE420 pKa = 4.21STYY423 pKa = 10.69TPFWSASPSSISSVAAVSSSAPDD446 pKa = 3.68TIVTVTVWEE455 pKa = 4.54TTDD458 pKa = 3.49SLLFPTGDD466 pKa = 3.36TVYY469 pKa = 10.78GGPSSNLPDD478 pKa = 5.26DD479 pKa = 4.63GVLTLTVTVTDD490 pKa = 4.79DD491 pKa = 3.57QPVWATDD498 pKa = 3.15VTTQTGYY505 pKa = 10.55GHH507 pKa = 6.79HH508 pKa = 5.77TTAANASGALPQPYY522 pKa = 10.15GSNTSSRR529 pKa = 11.84SSFDD533 pKa = 2.82PWLTTASGEE542 pKa = 4.32PYY544 pKa = 10.36GGPDD548 pKa = 3.41EE549 pKa = 4.47STVAVSEE556 pKa = 4.46VTFTQTVPLGPDD568 pKa = 2.87SSTVIVVTTEE578 pKa = 3.6VTVDD582 pKa = 3.24VTPTSSSSPTPSPSVIITPVVGTFTLSGAYY612 pKa = 7.56GTTIVLTYY620 pKa = 7.74TTEE623 pKa = 4.11LTITTALPPEE633 pKa = 4.51SSDD636 pKa = 3.74LSTGLVDD643 pKa = 3.89PTSLSPSFSGNFSSPSSSTSCSTSDD668 pKa = 3.45VLGLEE673 pKa = 4.35TSTFSQEE680 pKa = 3.99NPVPTEE686 pKa = 3.82VSVWPPPGPSSTSCSTTSSTSVLPLFTSASSSTTSCSNNSTTSSHH731 pKa = 6.55RR732 pKa = 11.84LAHH735 pKa = 5.74HH736 pKa = 6.83TSHH739 pKa = 7.05ASLGGSSTLGGYY751 pKa = 7.41VFPSSDD757 pKa = 3.03GAYY760 pKa = 9.79GGEE763 pKa = 3.82WSSDD767 pKa = 3.53SEE769 pKa = 4.39FSTVAPLPNSPSSTLTPVISSALPSINSNTILSTPEE805 pKa = 3.42VTLTDD810 pKa = 4.01GPTIPTWSFSSVNLPTISFVSSVSAITGSAEE841 pKa = 3.56ASAAPTAPASSLSTPIPVISTVPTLSSSVAPDD873 pKa = 3.46LTTSHH878 pKa = 7.03EE879 pKa = 4.54SPEE882 pKa = 4.14QTSSSISSSSGVSSSVLSHH901 pKa = 6.63LSSGTEE907 pKa = 3.84LSPSSTISHH916 pKa = 6.06TTTTSISGSPPPPFVPTSWVNSSAPSTLPTEE947 pKa = 4.5VSSTLNGTISPPATLTSSVPAEE969 pKa = 4.04LSSEE973 pKa = 3.73ISTTLNGTIPPVPLTISIVSVPTNVSSTLNGTITPTIPLTSNVVSTFTPSVLPTIPPMPLSIHH1036 pKa = 5.32QPKK1039 pKa = 10.59SSVFSLMTISRR1050 pKa = 11.84EE1051 pKa = 3.85VSPDD1055 pKa = 3.4YY1056 pKa = 10.59VAKK1059 pKa = 10.75CGAEE1063 pKa = 3.96EE1064 pKa = 4.13DD1065 pKa = 3.84RR1066 pKa = 11.84GSVVLTFDD1074 pKa = 5.87DD1075 pKa = 4.39IPPLSVDD1082 pKa = 3.05KK1083 pKa = 10.95SANGEE1088 pKa = 3.96IGPQPVPFPYY1098 pKa = 10.15HH1099 pKa = 6.77RR1100 pKa = 11.84FFFSPDD1106 pKa = 3.03LRR1108 pKa = 11.84VVSSSSVKK1116 pKa = 10.33YY1117 pKa = 10.45KK1118 pKa = 10.4PISGGQLLQHH1128 pKa = 5.91NHH1130 pKa = 4.93SHH1132 pKa = 6.74HH1133 pKa = 6.75SVAQLGLAWLRR1144 pKa = 11.84SNDD1147 pKa = 3.09CFRR1150 pKa = 11.84FNFRR1154 pKa = 11.84GISLGCDD1161 pKa = 2.86SVDD1164 pKa = 3.47APCVFTVTALNWDD1177 pKa = 3.86GVDD1180 pKa = 4.8DD1181 pKa = 4.44AVQGATTLEE1190 pKa = 4.33IPACPAATDD1199 pKa = 3.72CEE1201 pKa = 4.38LRR1203 pKa = 11.84RR1204 pKa = 11.84QILDD1208 pKa = 4.62PITASTFTNLTAVNITLSSPAQAQAWWADD1237 pKa = 4.0DD1238 pKa = 3.96IQISWADD1245 pKa = 3.63TGCNAAACRR1254 pKa = 11.84AKK1256 pKa = 10.92VPDD1259 pKa = 3.2RR1260 pKa = 11.84TMTNNSPKK1268 pKa = 10.29SFPGKK1273 pKa = 8.34PRR1275 pKa = 11.84RR1276 pKa = 11.84YY1277 pKa = 8.34MRR1279 pKa = 11.84WGSRR1283 pKa = 11.84QLHH1286 pKa = 5.67AA1287 pKa = 5.43
MM1 pKa = 7.86RR2 pKa = 11.84IWTSIACLSGMVSLATAAVVPRR24 pKa = 11.84VDD26 pKa = 4.75EE27 pKa = 4.43IALSSCTTSEE37 pKa = 3.91EE38 pKa = 4.52SVTVVTITVHH48 pKa = 5.83PQTATTITVTAGVSSPSSLSSALPFPFPGDD78 pKa = 3.27SSLGSEE84 pKa = 4.33STTCTDD90 pKa = 3.63EE91 pKa = 5.76DD92 pKa = 3.88SGSTDD97 pKa = 3.05VSIITVTPSLSWSFTFSTDD116 pKa = 3.26TTSSSADD123 pKa = 3.07AVTAGGPIQTVTVYY137 pKa = 10.48VPAPTPVTVTVGPPFSPSSASTVTSDD163 pKa = 3.48TEE165 pKa = 4.37CPVTITYY172 pKa = 7.78TLSPGPVFPSGFTGTLTLGGSKK194 pKa = 10.1PSSTPEE200 pKa = 3.52VSSAFDD206 pKa = 3.65PFSTFPEE213 pKa = 4.39SSTFGTPSSPILGPVTSSDD232 pKa = 3.4ADD234 pKa = 3.65SSDD237 pKa = 3.34VSVDD241 pKa = 3.17VVTYY245 pKa = 7.37TIPAQGTSSAQIGTVTVTSGAAQSTVSDD273 pKa = 3.83SVFTLTLSVFEE284 pKa = 4.93PSSPTYY290 pKa = 10.06NIPISSQPGTVTVTVTSAPDD310 pKa = 3.6DD311 pKa = 4.85DD312 pKa = 6.31GNDD315 pKa = 5.22DD316 pKa = 5.76DD317 pKa = 6.29DD318 pKa = 5.26DD319 pKa = 3.66WWTVWTNSWEE329 pKa = 4.39SISTITAIDD338 pKa = 3.72DD339 pKa = 3.7HH340 pKa = 8.2SSTVSSQDD348 pKa = 3.27ALSIWTSDD356 pKa = 2.91EE357 pKa = 3.72GATVVTVTEE366 pKa = 4.26IEE368 pKa = 4.35EE369 pKa = 4.2LTATPSSPGKK379 pKa = 9.23PWPTGYY385 pKa = 10.4DD386 pKa = 3.02SSGASVVVVTYY397 pKa = 7.17TIPGGSSTYY406 pKa = 9.56TVTYY410 pKa = 7.59TIPPLSTPSEE420 pKa = 4.21STYY423 pKa = 10.69TPFWSASPSSISSVAAVSSSAPDD446 pKa = 3.68TIVTVTVWEE455 pKa = 4.54TTDD458 pKa = 3.49SLLFPTGDD466 pKa = 3.36TVYY469 pKa = 10.78GGPSSNLPDD478 pKa = 5.26DD479 pKa = 4.63GVLTLTVTVTDD490 pKa = 4.79DD491 pKa = 3.57QPVWATDD498 pKa = 3.15VTTQTGYY505 pKa = 10.55GHH507 pKa = 6.79HH508 pKa = 5.77TTAANASGALPQPYY522 pKa = 10.15GSNTSSRR529 pKa = 11.84SSFDD533 pKa = 2.82PWLTTASGEE542 pKa = 4.32PYY544 pKa = 10.36GGPDD548 pKa = 3.41EE549 pKa = 4.47STVAVSEE556 pKa = 4.46VTFTQTVPLGPDD568 pKa = 2.87SSTVIVVTTEE578 pKa = 3.6VTVDD582 pKa = 3.24VTPTSSSSPTPSPSVIITPVVGTFTLSGAYY612 pKa = 7.56GTTIVLTYY620 pKa = 7.74TTEE623 pKa = 4.11LTITTALPPEE633 pKa = 4.51SSDD636 pKa = 3.74LSTGLVDD643 pKa = 3.89PTSLSPSFSGNFSSPSSSTSCSTSDD668 pKa = 3.45VLGLEE673 pKa = 4.35TSTFSQEE680 pKa = 3.99NPVPTEE686 pKa = 3.82VSVWPPPGPSSTSCSTTSSTSVLPLFTSASSSTTSCSNNSTTSSHH731 pKa = 6.55RR732 pKa = 11.84LAHH735 pKa = 5.74HH736 pKa = 6.83TSHH739 pKa = 7.05ASLGGSSTLGGYY751 pKa = 7.41VFPSSDD757 pKa = 3.03GAYY760 pKa = 9.79GGEE763 pKa = 3.82WSSDD767 pKa = 3.53SEE769 pKa = 4.39FSTVAPLPNSPSSTLTPVISSALPSINSNTILSTPEE805 pKa = 3.42VTLTDD810 pKa = 4.01GPTIPTWSFSSVNLPTISFVSSVSAITGSAEE841 pKa = 3.56ASAAPTAPASSLSTPIPVISTVPTLSSSVAPDD873 pKa = 3.46LTTSHH878 pKa = 7.03EE879 pKa = 4.54SPEE882 pKa = 4.14QTSSSISSSSGVSSSVLSHH901 pKa = 6.63LSSGTEE907 pKa = 3.84LSPSSTISHH916 pKa = 6.06TTTTSISGSPPPPFVPTSWVNSSAPSTLPTEE947 pKa = 4.5VSSTLNGTISPPATLTSSVPAEE969 pKa = 4.04LSSEE973 pKa = 3.73ISTTLNGTIPPVPLTISIVSVPTNVSSTLNGTITPTIPLTSNVVSTFTPSVLPTIPPMPLSIHH1036 pKa = 5.32QPKK1039 pKa = 10.59SSVFSLMTISRR1050 pKa = 11.84EE1051 pKa = 3.85VSPDD1055 pKa = 3.4YY1056 pKa = 10.59VAKK1059 pKa = 10.75CGAEE1063 pKa = 3.96EE1064 pKa = 4.13DD1065 pKa = 3.84RR1066 pKa = 11.84GSVVLTFDD1074 pKa = 5.87DD1075 pKa = 4.39IPPLSVDD1082 pKa = 3.05KK1083 pKa = 10.95SANGEE1088 pKa = 3.96IGPQPVPFPYY1098 pKa = 10.15HH1099 pKa = 6.77RR1100 pKa = 11.84FFFSPDD1106 pKa = 3.03LRR1108 pKa = 11.84VVSSSSVKK1116 pKa = 10.33YY1117 pKa = 10.45KK1118 pKa = 10.4PISGGQLLQHH1128 pKa = 5.91NHH1130 pKa = 4.93SHH1132 pKa = 6.74HH1133 pKa = 6.75SVAQLGLAWLRR1144 pKa = 11.84SNDD1147 pKa = 3.09CFRR1150 pKa = 11.84FNFRR1154 pKa = 11.84GISLGCDD1161 pKa = 2.86SVDD1164 pKa = 3.47APCVFTVTALNWDD1177 pKa = 3.86GVDD1180 pKa = 4.8DD1181 pKa = 4.44AVQGATTLEE1190 pKa = 4.33IPACPAATDD1199 pKa = 3.72CEE1201 pKa = 4.38LRR1203 pKa = 11.84RR1204 pKa = 11.84QILDD1208 pKa = 4.62PITASTFTNLTAVNITLSSPAQAQAWWADD1237 pKa = 4.0DD1238 pKa = 3.96IQISWADD1245 pKa = 3.63TGCNAAACRR1254 pKa = 11.84AKK1256 pKa = 10.92VPDD1259 pKa = 3.2RR1260 pKa = 11.84TMTNNSPKK1268 pKa = 10.29SFPGKK1273 pKa = 8.34PRR1275 pKa = 11.84RR1276 pKa = 11.84YY1277 pKa = 8.34MRR1279 pKa = 11.84WGSRR1283 pKa = 11.84QLHH1286 pKa = 5.67AA1287 pKa = 5.43
Molecular weight: 132.23 kDa
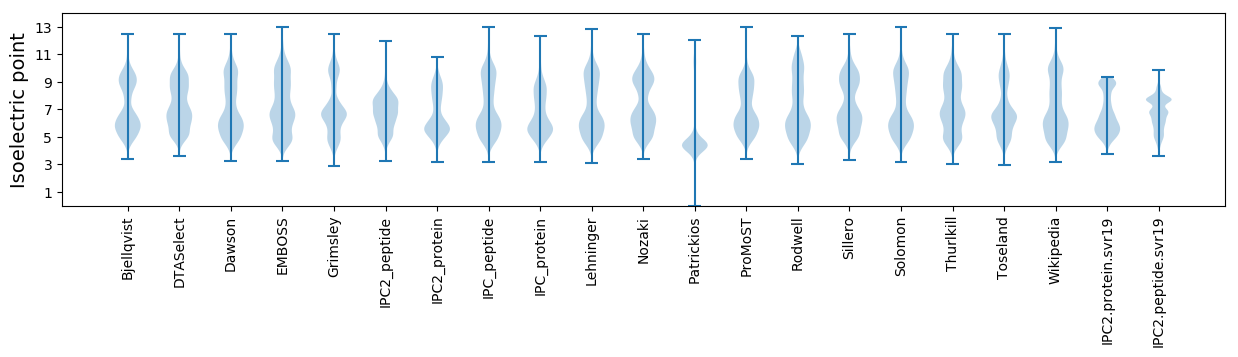
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G0S2Y1|G0S2Y1_CHATD Uncharacterized protein OS=Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) OX=759272 GN=CTHT_0018900 PE=4 SV=1
MM1 pKa = 7.36QDD3 pKa = 3.09EE4 pKa = 4.27EE5 pKa = 4.81LLRR8 pKa = 11.84KK9 pKa = 9.56PSQGKK14 pKa = 8.13LTSLLTDD21 pKa = 4.0WVHH24 pKa = 5.65GWQATALLLHH34 pKa = 7.28AIGICPMATPSVWYY48 pKa = 10.09SPHH51 pKa = 6.68GSLNTEE57 pKa = 3.49PMLRR61 pKa = 11.84ARR63 pKa = 11.84GIDD66 pKa = 3.36PHH68 pKa = 7.36NPSITNTAAAAARR81 pKa = 11.84IAAHH85 pKa = 5.94TALFTAAKK93 pKa = 8.83LHH95 pKa = 6.58HH96 pKa = 6.29GMPFAGAAARR106 pKa = 11.84SAIPQPIGSPRR117 pKa = 11.84TQVLKK122 pKa = 11.06LLLCRR127 pKa = 11.84SSMSGRR133 pKa = 11.84NPRR136 pKa = 11.84IRR138 pKa = 11.84SPYY141 pKa = 9.01FLSPTHH147 pKa = 5.54VV148 pKa = 3.13
MM1 pKa = 7.36QDD3 pKa = 3.09EE4 pKa = 4.27EE5 pKa = 4.81LLRR8 pKa = 11.84KK9 pKa = 9.56PSQGKK14 pKa = 8.13LTSLLTDD21 pKa = 4.0WVHH24 pKa = 5.65GWQATALLLHH34 pKa = 7.28AIGICPMATPSVWYY48 pKa = 10.09SPHH51 pKa = 6.68GSLNTEE57 pKa = 3.49PMLRR61 pKa = 11.84ARR63 pKa = 11.84GIDD66 pKa = 3.36PHH68 pKa = 7.36NPSITNTAAAAARR81 pKa = 11.84IAAHH85 pKa = 5.94TALFTAAKK93 pKa = 8.83LHH95 pKa = 6.58HH96 pKa = 6.29GMPFAGAAARR106 pKa = 11.84SAIPQPIGSPRR117 pKa = 11.84TQVLKK122 pKa = 11.06LLLCRR127 pKa = 11.84SSMSGRR133 pKa = 11.84NPRR136 pKa = 11.84IRR138 pKa = 11.84SPYY141 pKa = 9.01FLSPTHH147 pKa = 5.54VV148 pKa = 3.13
Molecular weight: 15.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3924665 |
22 |
8081 |
546.2 |
60.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.039 ± 0.029 | 1.086 ± 0.01 |
5.489 ± 0.02 | 6.696 ± 0.047 |
3.365 ± 0.018 | 6.853 ± 0.029 |
2.3 ± 0.012 | 4.512 ± 0.018 |
5.007 ± 0.028 | 8.664 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.019 ± 0.011 | 3.494 ± 0.016 |
6.867 ± 0.038 | 4.194 ± 0.029 |
6.505 ± 0.028 | 7.993 ± 0.034 |
5.901 ± 0.02 | 6.082 ± 0.025 |
1.368 ± 0.011 | 2.563 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
