
Bacillus lacisalsi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
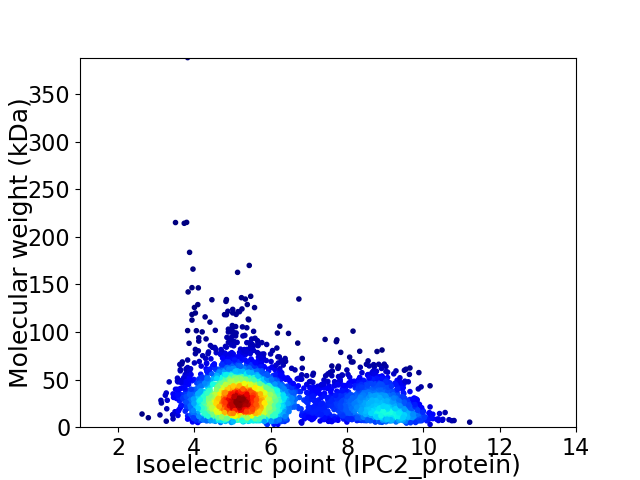
Virtual 2D-PAGE plot for 3874 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2W0H9X8|A0A2W0H9X8_9BACI 5-oxoprolinase subunit A OS=Bacillus lacisalsi OX=2045244 GN=pxpA PE=3 SV=1
MM1 pKa = 7.54KK2 pKa = 10.39KK3 pKa = 10.0LLAGAGVLAAAAVIAGGIYY22 pKa = 10.67AFTNDD27 pKa = 3.89SAPSAGPDD35 pKa = 3.24YY36 pKa = 10.16EE37 pKa = 4.57NPVFEE42 pKa = 4.96PVIADD47 pKa = 3.49PSVIRR52 pKa = 11.84GEE54 pKa = 4.15DD55 pKa = 3.84GYY57 pKa = 11.22FYY59 pKa = 10.91AYY61 pKa = 8.98GTEE64 pKa = 4.43DD65 pKa = 2.87NWGDD69 pKa = 3.54GMGSRR74 pKa = 11.84VVPIVRR80 pKa = 11.84SQNLTDD86 pKa = 3.26WDD88 pKa = 4.32YY89 pKa = 11.92VGEE92 pKa = 4.28AFQTKK97 pKa = 9.77PDD99 pKa = 3.32WKK101 pKa = 10.17EE102 pKa = 3.99DD103 pKa = 3.49GGIWAPDD110 pKa = 2.75IVYY113 pKa = 10.45FNDD116 pKa = 2.98QYY118 pKa = 11.73YY119 pKa = 10.75LYY121 pKa = 9.95YY122 pKa = 10.55SQSTWGDD129 pKa = 3.53PNPAIGVAVADD140 pKa = 4.75DD141 pKa = 3.85PAGPFDD147 pKa = 4.51DD148 pKa = 3.97QGKK151 pKa = 9.39LFDD154 pKa = 4.13SEE156 pKa = 4.78EE157 pKa = 4.06IGVPNSIDD165 pKa = 3.16PQLFIKK171 pKa = 10.58EE172 pKa = 4.51DD173 pKa = 3.43GTPYY177 pKa = 10.71LFWGSWYY184 pKa = 10.02GIWGIEE190 pKa = 4.09ISDD193 pKa = 4.63DD194 pKa = 3.31GLDD197 pKa = 3.63YY198 pKa = 11.34VGEE201 pKa = 4.21KK202 pKa = 10.33FQIAGTDD209 pKa = 3.8FEE211 pKa = 4.71APYY214 pKa = 10.24IIEE217 pKa = 4.49RR218 pKa = 11.84DD219 pKa = 3.37DD220 pKa = 3.34YY221 pKa = 11.46FYY223 pKa = 11.39FFGSKK228 pKa = 10.43GSCCEE233 pKa = 4.12GQFSEE238 pKa = 4.29YY239 pKa = 9.69RR240 pKa = 11.84VAVGRR245 pKa = 11.84SEE247 pKa = 5.41SFDD250 pKa = 3.54GPYY253 pKa = 10.21LDD255 pKa = 5.33KK256 pKa = 11.32DD257 pKa = 3.84GNEE260 pKa = 4.66LNDD263 pKa = 3.4SSGTVILSGGEE274 pKa = 4.02GFAGPGHH281 pKa = 6.16NAVITDD287 pKa = 4.22DD288 pKa = 5.39AGQDD292 pKa = 2.89WMIYY296 pKa = 9.75HH297 pKa = 7.74AIDD300 pKa = 5.44KK301 pKa = 6.7EE302 pKa = 4.8TPWIGSGVTRR312 pKa = 11.84RR313 pKa = 11.84PLMLDD318 pKa = 3.6RR319 pKa = 11.84IIWEE323 pKa = 4.65DD324 pKa = 3.27GWPQIEE330 pKa = 4.28GGVPGEE336 pKa = 4.43GSQEE340 pKa = 3.86GPAIEE345 pKa = 4.01QQ346 pKa = 3.25
MM1 pKa = 7.54KK2 pKa = 10.39KK3 pKa = 10.0LLAGAGVLAAAAVIAGGIYY22 pKa = 10.67AFTNDD27 pKa = 3.89SAPSAGPDD35 pKa = 3.24YY36 pKa = 10.16EE37 pKa = 4.57NPVFEE42 pKa = 4.96PVIADD47 pKa = 3.49PSVIRR52 pKa = 11.84GEE54 pKa = 4.15DD55 pKa = 3.84GYY57 pKa = 11.22FYY59 pKa = 10.91AYY61 pKa = 8.98GTEE64 pKa = 4.43DD65 pKa = 2.87NWGDD69 pKa = 3.54GMGSRR74 pKa = 11.84VVPIVRR80 pKa = 11.84SQNLTDD86 pKa = 3.26WDD88 pKa = 4.32YY89 pKa = 11.92VGEE92 pKa = 4.28AFQTKK97 pKa = 9.77PDD99 pKa = 3.32WKK101 pKa = 10.17EE102 pKa = 3.99DD103 pKa = 3.49GGIWAPDD110 pKa = 2.75IVYY113 pKa = 10.45FNDD116 pKa = 2.98QYY118 pKa = 11.73YY119 pKa = 10.75LYY121 pKa = 9.95YY122 pKa = 10.55SQSTWGDD129 pKa = 3.53PNPAIGVAVADD140 pKa = 4.75DD141 pKa = 3.85PAGPFDD147 pKa = 4.51DD148 pKa = 3.97QGKK151 pKa = 9.39LFDD154 pKa = 4.13SEE156 pKa = 4.78EE157 pKa = 4.06IGVPNSIDD165 pKa = 3.16PQLFIKK171 pKa = 10.58EE172 pKa = 4.51DD173 pKa = 3.43GTPYY177 pKa = 10.71LFWGSWYY184 pKa = 10.02GIWGIEE190 pKa = 4.09ISDD193 pKa = 4.63DD194 pKa = 3.31GLDD197 pKa = 3.63YY198 pKa = 11.34VGEE201 pKa = 4.21KK202 pKa = 10.33FQIAGTDD209 pKa = 3.8FEE211 pKa = 4.71APYY214 pKa = 10.24IIEE217 pKa = 4.49RR218 pKa = 11.84DD219 pKa = 3.37DD220 pKa = 3.34YY221 pKa = 11.46FYY223 pKa = 11.39FFGSKK228 pKa = 10.43GSCCEE233 pKa = 4.12GQFSEE238 pKa = 4.29YY239 pKa = 9.69RR240 pKa = 11.84VAVGRR245 pKa = 11.84SEE247 pKa = 5.41SFDD250 pKa = 3.54GPYY253 pKa = 10.21LDD255 pKa = 5.33KK256 pKa = 11.32DD257 pKa = 3.84GNEE260 pKa = 4.66LNDD263 pKa = 3.4SSGTVILSGGEE274 pKa = 4.02GFAGPGHH281 pKa = 6.16NAVITDD287 pKa = 4.22DD288 pKa = 5.39AGQDD292 pKa = 2.89WMIYY296 pKa = 9.75HH297 pKa = 7.74AIDD300 pKa = 5.44KK301 pKa = 6.7EE302 pKa = 4.8TPWIGSGVTRR312 pKa = 11.84RR313 pKa = 11.84PLMLDD318 pKa = 3.6RR319 pKa = 11.84IIWEE323 pKa = 4.65DD324 pKa = 3.27GWPQIEE330 pKa = 4.28GGVPGEE336 pKa = 4.43GSQEE340 pKa = 3.86GPAIEE345 pKa = 4.01QQ346 pKa = 3.25
Molecular weight: 37.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2W0H5S2|A0A2W0H5S2_9BACI NERD domain-containing protein OS=Bacillus lacisalsi OX=2045244 GN=CR205_18490 PE=4 SV=1
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.18RR13 pKa = 11.84KK14 pKa = 8.28KK15 pKa = 8.46VHH17 pKa = 5.5GFRR20 pKa = 11.84TRR22 pKa = 11.84MSTKK26 pKa = 9.32NGRR29 pKa = 11.84RR30 pKa = 11.84VLASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.73GRR40 pKa = 11.84KK41 pKa = 8.76VLSAA45 pKa = 4.05
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.18RR13 pKa = 11.84KK14 pKa = 8.28KK15 pKa = 8.46VHH17 pKa = 5.5GFRR20 pKa = 11.84TRR22 pKa = 11.84MSTKK26 pKa = 9.32NGRR29 pKa = 11.84RR30 pKa = 11.84VLASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.73GRR40 pKa = 11.84KK41 pKa = 8.76VLSAA45 pKa = 4.05
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1131585 |
26 |
3595 |
292.1 |
32.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.968 ± 0.042 | 0.7 ± 0.013 |
5.628 ± 0.043 | 8.05 ± 0.057 |
4.477 ± 0.03 | 7.62 ± 0.042 |
2.186 ± 0.017 | 6.426 ± 0.037 |
5.45 ± 0.044 | 9.485 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.883 ± 0.019 | 3.635 ± 0.03 |
3.902 ± 0.026 | 3.358 ± 0.026 |
5.072 ± 0.034 | 5.847 ± 0.03 |
5.57 ± 0.036 | 7.36 ± 0.032 |
1.105 ± 0.016 | 3.279 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
