
Methanosarcina virus MetMV
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
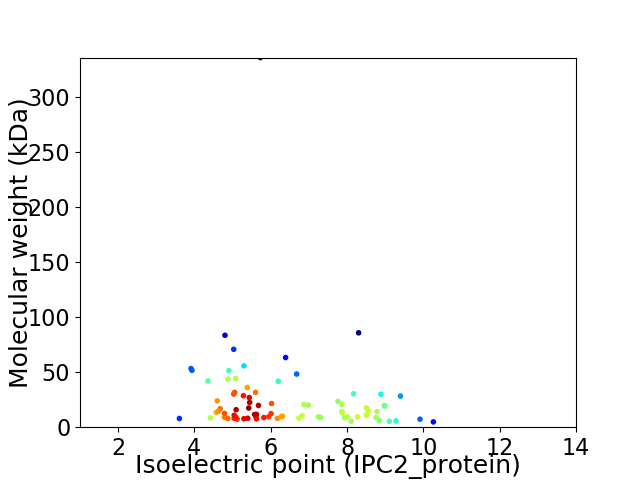
Virtual 2D-PAGE plot for 79 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A5K7NIT2|A0A5K7NIT2_9VIRU Uncharacterized protein OS=Methanosarcina virus MetMV OX=2488574 PE=4 SV=1
MM1 pKa = 7.34SKK3 pKa = 10.52VVLFTRR9 pKa = 11.84GNPWDD14 pKa = 3.78PSGIGSGTTSVLNTVSQTFSVPVTPQMKK42 pKa = 9.68FWLAGNGVSSVLITSARR59 pKa = 11.84LIQGTKK65 pKa = 10.23NIPVTVNGAASFYY78 pKa = 10.77IPATGKK84 pKa = 8.99WSDD87 pKa = 3.65YY88 pKa = 11.42ASISDD93 pKa = 4.1FSLPGNVTLTVTFSGTMNIATFLDD117 pKa = 3.66NSNPGDD123 pKa = 3.79PVVVSATEE131 pKa = 3.9IPSPEE136 pKa = 4.21TPSLAGEE143 pKa = 3.86ILFDD147 pKa = 4.02VEE149 pKa = 4.56FEE151 pKa = 4.34GSFVEE156 pKa = 4.69SVSGLMPSTIQNCTIEE172 pKa = 4.21TGNVLFGSGSCFCNGSEE189 pKa = 4.5SSPAVLIYY197 pKa = 9.56STNSTFVNRR206 pKa = 11.84CTEE209 pKa = 4.26DD210 pKa = 3.4NFTMSAFYY218 pKa = 10.57KK219 pKa = 9.74PLEE222 pKa = 4.35PFGEE226 pKa = 4.52YY227 pKa = 9.67PAGSRR232 pKa = 11.84IHH234 pKa = 5.84EE235 pKa = 3.77AVVFVKK241 pKa = 10.7VSGGYY246 pKa = 9.55NKK248 pKa = 9.9YY249 pKa = 10.58HH250 pKa = 6.31YY251 pKa = 10.91FNGTNFNDD259 pKa = 3.77PPFEE263 pKa = 4.29FEE265 pKa = 3.92SDD267 pKa = 3.3ASVEE271 pKa = 4.19THH273 pKa = 6.86LLNLMIGGAIQIIFDD288 pKa = 4.44GYY290 pKa = 10.47QIIKK294 pKa = 9.05GAVWTEE300 pKa = 3.93EE301 pKa = 4.15FTPPSQPFIFSSDD314 pKa = 3.28YY315 pKa = 11.51GLIEE319 pKa = 4.06LTSNSSITLTTNPAPGDD336 pKa = 3.84TIKK339 pKa = 11.21LKK341 pKa = 10.93GGTLGDD347 pKa = 3.54VTLTFVNYY355 pKa = 9.58YY356 pKa = 9.71PRR358 pKa = 11.84NQYY361 pKa = 8.92EE362 pKa = 4.21VQIGATVNEE371 pKa = 4.3TAANLTLAINRR382 pKa = 11.84IEE384 pKa = 4.17GAEE387 pKa = 4.11FTATATDD394 pKa = 3.51NVVDD398 pKa = 5.29IIAYY402 pKa = 9.84SPTFSTEE409 pKa = 3.48KK410 pKa = 9.97TGDD413 pKa = 3.97GITLSGVSSTLDD425 pKa = 3.29GLITEE430 pKa = 5.73DD431 pKa = 4.1ISVDD435 pKa = 3.53DD436 pKa = 4.68SFTVTDD442 pKa = 3.91GGKK445 pKa = 10.3AIQQNVSVTEE455 pKa = 4.82AIDD458 pKa = 3.71GLIDD462 pKa = 4.03GLSEE466 pKa = 4.47NINITDD472 pKa = 3.8YY473 pKa = 11.24FDD475 pKa = 3.68TSMDD479 pKa = 3.63GLDD482 pKa = 3.48EE483 pKa = 4.4SVLVSTSFVGLIDD496 pKa = 3.79VMGAVV501 pKa = 3.88
MM1 pKa = 7.34SKK3 pKa = 10.52VVLFTRR9 pKa = 11.84GNPWDD14 pKa = 3.78PSGIGSGTTSVLNTVSQTFSVPVTPQMKK42 pKa = 9.68FWLAGNGVSSVLITSARR59 pKa = 11.84LIQGTKK65 pKa = 10.23NIPVTVNGAASFYY78 pKa = 10.77IPATGKK84 pKa = 8.99WSDD87 pKa = 3.65YY88 pKa = 11.42ASISDD93 pKa = 4.1FSLPGNVTLTVTFSGTMNIATFLDD117 pKa = 3.66NSNPGDD123 pKa = 3.79PVVVSATEE131 pKa = 3.9IPSPEE136 pKa = 4.21TPSLAGEE143 pKa = 3.86ILFDD147 pKa = 4.02VEE149 pKa = 4.56FEE151 pKa = 4.34GSFVEE156 pKa = 4.69SVSGLMPSTIQNCTIEE172 pKa = 4.21TGNVLFGSGSCFCNGSEE189 pKa = 4.5SSPAVLIYY197 pKa = 9.56STNSTFVNRR206 pKa = 11.84CTEE209 pKa = 4.26DD210 pKa = 3.4NFTMSAFYY218 pKa = 10.57KK219 pKa = 9.74PLEE222 pKa = 4.35PFGEE226 pKa = 4.52YY227 pKa = 9.67PAGSRR232 pKa = 11.84IHH234 pKa = 5.84EE235 pKa = 3.77AVVFVKK241 pKa = 10.7VSGGYY246 pKa = 9.55NKK248 pKa = 9.9YY249 pKa = 10.58HH250 pKa = 6.31YY251 pKa = 10.91FNGTNFNDD259 pKa = 3.77PPFEE263 pKa = 4.29FEE265 pKa = 3.92SDD267 pKa = 3.3ASVEE271 pKa = 4.19THH273 pKa = 6.86LLNLMIGGAIQIIFDD288 pKa = 4.44GYY290 pKa = 10.47QIIKK294 pKa = 9.05GAVWTEE300 pKa = 3.93EE301 pKa = 4.15FTPPSQPFIFSSDD314 pKa = 3.28YY315 pKa = 11.51GLIEE319 pKa = 4.06LTSNSSITLTTNPAPGDD336 pKa = 3.84TIKK339 pKa = 11.21LKK341 pKa = 10.93GGTLGDD347 pKa = 3.54VTLTFVNYY355 pKa = 9.58YY356 pKa = 9.71PRR358 pKa = 11.84NQYY361 pKa = 8.92EE362 pKa = 4.21VQIGATVNEE371 pKa = 4.3TAANLTLAINRR382 pKa = 11.84IEE384 pKa = 4.17GAEE387 pKa = 4.11FTATATDD394 pKa = 3.51NVVDD398 pKa = 5.29IIAYY402 pKa = 9.84SPTFSTEE409 pKa = 3.48KK410 pKa = 9.97TGDD413 pKa = 3.97GITLSGVSSTLDD425 pKa = 3.29GLITEE430 pKa = 5.73DD431 pKa = 4.1ISVDD435 pKa = 3.53DD436 pKa = 4.68SFTVTDD442 pKa = 3.91GGKK445 pKa = 10.3AIQQNVSVTEE455 pKa = 4.82AIDD458 pKa = 3.71GLIDD462 pKa = 4.03GLSEE466 pKa = 4.47NINITDD472 pKa = 3.8YY473 pKa = 11.24FDD475 pKa = 3.68TSMDD479 pKa = 3.63GLDD482 pKa = 3.48EE483 pKa = 4.4SVLVSTSFVGLIDD496 pKa = 3.79VMGAVV501 pKa = 3.88
Molecular weight: 53.26 kDa
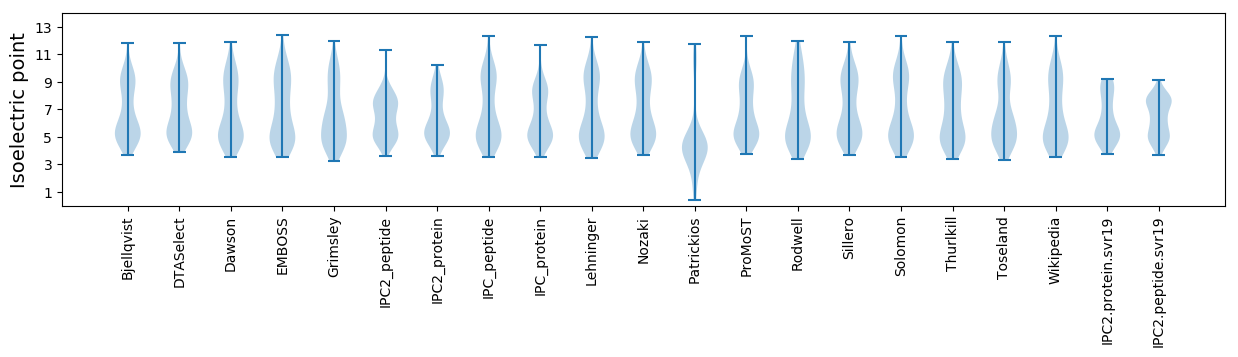
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5K7NMS3|A0A5K7NMS3_9VIRU Uncharacterized protein OS=Methanosarcina virus MetMV OX=2488574 PE=4 SV=1
MM1 pKa = 7.8RR2 pKa = 11.84KK3 pKa = 9.5KK4 pKa = 10.71LLEE7 pKa = 4.13KK8 pKa = 10.46LKK10 pKa = 10.84KK11 pKa = 9.84DD12 pKa = 3.45VKK14 pKa = 10.56KK15 pKa = 10.63YY16 pKa = 10.96GVMEE20 pKa = 3.79MSRR23 pKa = 11.84RR24 pKa = 11.84LRR26 pKa = 11.84WPYY29 pKa = 9.32MPLYY33 pKa = 10.59RR34 pKa = 11.84LINGKK39 pKa = 8.84NAGHH43 pKa = 6.61INLWDD48 pKa = 4.9RR49 pKa = 11.84IEE51 pKa = 4.79AYY53 pKa = 10.1YY54 pKa = 10.85KK55 pKa = 10.2KK56 pKa = 10.59AKK58 pKa = 9.65KK59 pKa = 9.96
MM1 pKa = 7.8RR2 pKa = 11.84KK3 pKa = 9.5KK4 pKa = 10.71LLEE7 pKa = 4.13KK8 pKa = 10.46LKK10 pKa = 10.84KK11 pKa = 9.84DD12 pKa = 3.45VKK14 pKa = 10.56KK15 pKa = 10.63YY16 pKa = 10.96GVMEE20 pKa = 3.79MSRR23 pKa = 11.84RR24 pKa = 11.84LRR26 pKa = 11.84WPYY29 pKa = 9.32MPLYY33 pKa = 10.59RR34 pKa = 11.84LINGKK39 pKa = 8.84NAGHH43 pKa = 6.61INLWDD48 pKa = 4.9RR49 pKa = 11.84IEE51 pKa = 4.79AYY53 pKa = 10.1YY54 pKa = 10.85KK55 pKa = 10.2KK56 pKa = 10.59AKK58 pKa = 9.65KK59 pKa = 9.96
Molecular weight: 7.3 kDa
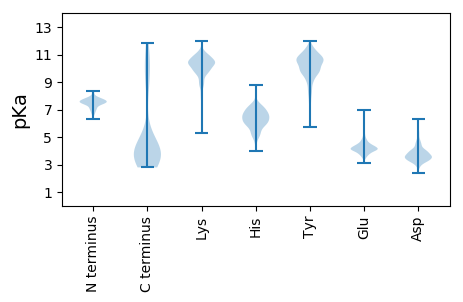
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
18061 |
39 |
3002 |
228.6 |
25.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.084 ± 0.409 | 1.44 ± 0.239 |
5.919 ± 0.177 | 7.131 ± 0.434 |
3.455 ± 0.15 | 7.026 ± 0.367 |
1.55 ± 0.125 | 7.026 ± 0.238 |
7.691 ± 0.426 | 7.663 ± 0.224 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.215 ± 0.131 | 5.52 ± 0.331 |
3.754 ± 0.263 | 3.488 ± 0.189 |
4.319 ± 0.246 | 6.03 ± 0.333 |
6.262 ± 0.391 | 5.825 ± 0.221 |
1.528 ± 0.123 | 4.075 ± 0.234 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
