
Clostridium sp. CAG:440
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
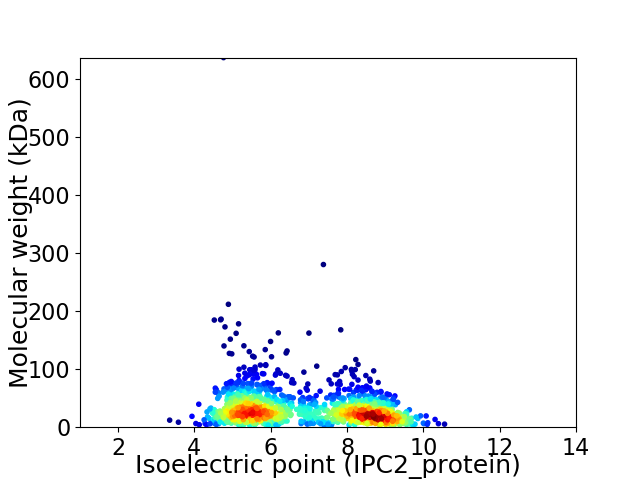
Virtual 2D-PAGE plot for 1420 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7GDC8|R7GDC8_9CLOT Uncharacterized protein OS=Clostridium sp. CAG:440 OX=1262807 GN=BN658_00694 PE=4 SV=1
MM1 pKa = 7.32IKK3 pKa = 10.31EE4 pKa = 3.94NNEE7 pKa = 3.64EE8 pKa = 4.01FFKK11 pKa = 11.07NICQKK16 pKa = 11.15YY17 pKa = 8.09IDD19 pKa = 4.02TMSKK23 pKa = 10.68EE24 pKa = 4.19MLLEE28 pKa = 4.53ADD30 pKa = 3.98NKK32 pKa = 10.2KK33 pKa = 9.83ILYY36 pKa = 9.87IFIEE40 pKa = 4.25DD41 pKa = 3.86EE42 pKa = 4.28YY43 pKa = 11.21IEE45 pKa = 4.67LKK47 pKa = 10.71EE48 pKa = 4.26KK49 pKa = 11.02GSINIQNYY57 pKa = 9.68KK58 pKa = 10.73DD59 pKa = 3.8YY60 pKa = 11.29LDD62 pKa = 4.48GDD64 pKa = 4.12FTVYY68 pKa = 10.39QYY70 pKa = 11.83DD71 pKa = 3.94SYY73 pKa = 11.75QEE75 pKa = 4.22LYY77 pKa = 10.53KK78 pKa = 10.24VTIKK82 pKa = 11.13DD83 pKa = 3.55EE84 pKa = 4.04ILNDD88 pKa = 3.64IEE90 pKa = 5.48DD91 pKa = 4.47LGLFDD96 pKa = 6.98DD97 pKa = 4.18NNQWNFYY104 pKa = 8.82ITFEE108 pKa = 3.89EE109 pKa = 4.54LKK111 pKa = 10.41RR112 pKa = 11.84LEE114 pKa = 4.08YY115 pKa = 10.51GYY117 pKa = 10.71KK118 pKa = 9.17VQYY121 pKa = 10.92NYY123 pKa = 10.51TLLEE127 pKa = 3.97KK128 pKa = 10.87YY129 pKa = 10.14IIEE132 pKa = 4.3NEE134 pKa = 3.93EE135 pKa = 3.62DD136 pKa = 4.03FYY138 pKa = 10.98TYY140 pKa = 10.72FSLEE144 pKa = 3.69QLNEE148 pKa = 4.15FEE150 pKa = 4.98EE151 pKa = 4.69SLHH154 pKa = 7.02LYY156 pKa = 10.8YY157 pKa = 10.27EE158 pKa = 4.32TDD160 pKa = 5.45DD161 pKa = 3.57IDD163 pKa = 3.86TNKK166 pKa = 9.92WGEE169 pKa = 4.36LYY171 pKa = 10.87SKK173 pKa = 10.55SNKK176 pKa = 9.29YY177 pKa = 10.35FNNNIIALSEE187 pKa = 4.06GMISYY192 pKa = 11.11YY193 pKa = 10.92DD194 pKa = 3.76LLQDD198 pKa = 4.02YY199 pKa = 9.86KK200 pKa = 11.03INYY203 pKa = 5.69QQYY206 pKa = 10.08YY207 pKa = 9.9GLSLDD212 pKa = 3.72KK213 pKa = 10.92VIEE216 pKa = 4.08YY217 pKa = 10.19FRR219 pKa = 11.84EE220 pKa = 3.81NAIVNLMDD228 pKa = 3.75YY229 pKa = 11.12GSDD232 pKa = 3.32RR233 pKa = 11.84DD234 pKa = 3.79EE235 pKa = 4.97GLHH238 pKa = 5.7QLSSMYY244 pKa = 10.48KK245 pKa = 10.09EE246 pKa = 4.13IMDD249 pKa = 4.0NLGIKK254 pKa = 9.66YY255 pKa = 10.34SNVYY259 pKa = 9.27TEE261 pKa = 4.74DD262 pKa = 3.65VSDD265 pKa = 3.82GKK267 pKa = 11.24YY268 pKa = 8.99LTTITFEE275 pKa = 4.41DD276 pKa = 4.87DD277 pKa = 3.22YY278 pKa = 11.25SIEE281 pKa = 4.18VDD283 pKa = 3.32TSAWNGIEE291 pKa = 4.04VVTEE295 pKa = 4.25NIEE298 pKa = 4.43SIYY301 pKa = 10.8EE302 pKa = 4.03EE303 pKa = 4.48YY304 pKa = 10.57EE305 pKa = 4.01KK306 pKa = 10.88QSEE309 pKa = 4.22KK310 pKa = 10.37TKK312 pKa = 10.63QNEE315 pKa = 4.14VKK317 pKa = 10.55NEE319 pKa = 3.75KK320 pKa = 10.19EE321 pKa = 3.75FEE323 pKa = 4.04YY324 pKa = 11.1DD325 pKa = 3.64FNN327 pKa = 4.76
MM1 pKa = 7.32IKK3 pKa = 10.31EE4 pKa = 3.94NNEE7 pKa = 3.64EE8 pKa = 4.01FFKK11 pKa = 11.07NICQKK16 pKa = 11.15YY17 pKa = 8.09IDD19 pKa = 4.02TMSKK23 pKa = 10.68EE24 pKa = 4.19MLLEE28 pKa = 4.53ADD30 pKa = 3.98NKK32 pKa = 10.2KK33 pKa = 9.83ILYY36 pKa = 9.87IFIEE40 pKa = 4.25DD41 pKa = 3.86EE42 pKa = 4.28YY43 pKa = 11.21IEE45 pKa = 4.67LKK47 pKa = 10.71EE48 pKa = 4.26KK49 pKa = 11.02GSINIQNYY57 pKa = 9.68KK58 pKa = 10.73DD59 pKa = 3.8YY60 pKa = 11.29LDD62 pKa = 4.48GDD64 pKa = 4.12FTVYY68 pKa = 10.39QYY70 pKa = 11.83DD71 pKa = 3.94SYY73 pKa = 11.75QEE75 pKa = 4.22LYY77 pKa = 10.53KK78 pKa = 10.24VTIKK82 pKa = 11.13DD83 pKa = 3.55EE84 pKa = 4.04ILNDD88 pKa = 3.64IEE90 pKa = 5.48DD91 pKa = 4.47LGLFDD96 pKa = 6.98DD97 pKa = 4.18NNQWNFYY104 pKa = 8.82ITFEE108 pKa = 3.89EE109 pKa = 4.54LKK111 pKa = 10.41RR112 pKa = 11.84LEE114 pKa = 4.08YY115 pKa = 10.51GYY117 pKa = 10.71KK118 pKa = 9.17VQYY121 pKa = 10.92NYY123 pKa = 10.51TLLEE127 pKa = 3.97KK128 pKa = 10.87YY129 pKa = 10.14IIEE132 pKa = 4.3NEE134 pKa = 3.93EE135 pKa = 3.62DD136 pKa = 4.03FYY138 pKa = 10.98TYY140 pKa = 10.72FSLEE144 pKa = 3.69QLNEE148 pKa = 4.15FEE150 pKa = 4.98EE151 pKa = 4.69SLHH154 pKa = 7.02LYY156 pKa = 10.8YY157 pKa = 10.27EE158 pKa = 4.32TDD160 pKa = 5.45DD161 pKa = 3.57IDD163 pKa = 3.86TNKK166 pKa = 9.92WGEE169 pKa = 4.36LYY171 pKa = 10.87SKK173 pKa = 10.55SNKK176 pKa = 9.29YY177 pKa = 10.35FNNNIIALSEE187 pKa = 4.06GMISYY192 pKa = 11.11YY193 pKa = 10.92DD194 pKa = 3.76LLQDD198 pKa = 4.02YY199 pKa = 9.86KK200 pKa = 11.03INYY203 pKa = 5.69QQYY206 pKa = 10.08YY207 pKa = 9.9GLSLDD212 pKa = 3.72KK213 pKa = 10.92VIEE216 pKa = 4.08YY217 pKa = 10.19FRR219 pKa = 11.84EE220 pKa = 3.81NAIVNLMDD228 pKa = 3.75YY229 pKa = 11.12GSDD232 pKa = 3.32RR233 pKa = 11.84DD234 pKa = 3.79EE235 pKa = 4.97GLHH238 pKa = 5.7QLSSMYY244 pKa = 10.48KK245 pKa = 10.09EE246 pKa = 4.13IMDD249 pKa = 4.0NLGIKK254 pKa = 9.66YY255 pKa = 10.34SNVYY259 pKa = 9.27TEE261 pKa = 4.74DD262 pKa = 3.65VSDD265 pKa = 3.82GKK267 pKa = 11.24YY268 pKa = 8.99LTTITFEE275 pKa = 4.41DD276 pKa = 4.87DD277 pKa = 3.22YY278 pKa = 11.25SIEE281 pKa = 4.18VDD283 pKa = 3.32TSAWNGIEE291 pKa = 4.04VVTEE295 pKa = 4.25NIEE298 pKa = 4.43SIYY301 pKa = 10.8EE302 pKa = 4.03EE303 pKa = 4.48YY304 pKa = 10.57EE305 pKa = 4.01KK306 pKa = 10.88QSEE309 pKa = 4.22KK310 pKa = 10.37TKK312 pKa = 10.63QNEE315 pKa = 4.14VKK317 pKa = 10.55NEE319 pKa = 3.75KK320 pKa = 10.19EE321 pKa = 3.75FEE323 pKa = 4.04YY324 pKa = 11.1DD325 pKa = 3.64FNN327 pKa = 4.76
Molecular weight: 39.61 kDa
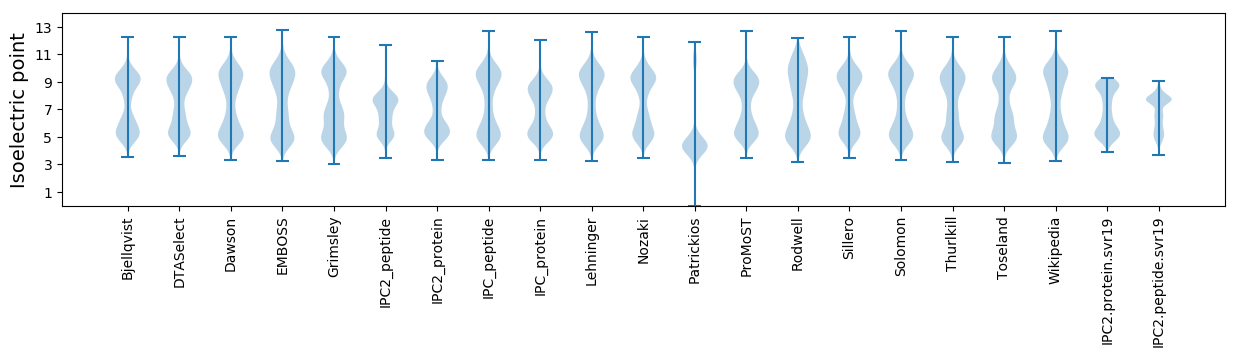
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7GCC1|R7GCC1_9CLOT Sporulation stage III protein AA OS=Clostridium sp. CAG:440 OX=1262807 GN=BN658_00370 PE=4 SV=1
MM1 pKa = 7.79AKK3 pKa = 9.41TNNNIVFYY11 pKa = 9.09GTGRR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 10.29SSIARR22 pKa = 11.84VRR24 pKa = 11.84LVEE27 pKa = 4.01GSGKK31 pKa = 8.35ITINGKK37 pKa = 10.22DD38 pKa = 2.95IDD40 pKa = 4.17EE41 pKa = 4.57FFGLEE46 pKa = 3.93TLKK49 pKa = 11.23VIVRR53 pKa = 11.84QPLTVTNTTAKK64 pKa = 10.26YY65 pKa = 10.7DD66 pKa = 3.73VICTVKK72 pKa = 10.79GGGFTGQAGAIRR84 pKa = 11.84HH85 pKa = 6.08GIARR89 pKa = 11.84ALNEE93 pKa = 4.19ANSEE97 pKa = 4.02YY98 pKa = 10.53RR99 pKa = 11.84PSLKK103 pKa = 10.1TNGFLTRR110 pKa = 11.84DD111 pKa = 3.3PRR113 pKa = 11.84MKK115 pKa = 10.18EE116 pKa = 3.54RR117 pKa = 11.84KK118 pKa = 9.62KK119 pKa = 11.12YY120 pKa = 9.99GLKK123 pKa = 10.15KK124 pKa = 10.22ARR126 pKa = 11.84KK127 pKa = 8.46APQFSKK133 pKa = 10.93RR134 pKa = 3.46
MM1 pKa = 7.79AKK3 pKa = 9.41TNNNIVFYY11 pKa = 9.09GTGRR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 10.29SSIARR22 pKa = 11.84VRR24 pKa = 11.84LVEE27 pKa = 4.01GSGKK31 pKa = 8.35ITINGKK37 pKa = 10.22DD38 pKa = 2.95IDD40 pKa = 4.17EE41 pKa = 4.57FFGLEE46 pKa = 3.93TLKK49 pKa = 11.23VIVRR53 pKa = 11.84QPLTVTNTTAKK64 pKa = 10.26YY65 pKa = 10.7DD66 pKa = 3.73VICTVKK72 pKa = 10.79GGGFTGQAGAIRR84 pKa = 11.84HH85 pKa = 6.08GIARR89 pKa = 11.84ALNEE93 pKa = 4.19ANSEE97 pKa = 4.02YY98 pKa = 10.53RR99 pKa = 11.84PSLKK103 pKa = 10.1TNGFLTRR110 pKa = 11.84DD111 pKa = 3.3PRR113 pKa = 11.84MKK115 pKa = 10.18EE116 pKa = 3.54RR117 pKa = 11.84KK118 pKa = 9.62KK119 pKa = 11.12YY120 pKa = 9.99GLKK123 pKa = 10.15KK124 pKa = 10.22ARR126 pKa = 11.84KK127 pKa = 8.46APQFSKK133 pKa = 10.93RR134 pKa = 3.46
Molecular weight: 14.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
418409 |
29 |
5824 |
294.7 |
33.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.954 ± 0.06 | 1.089 ± 0.029 |
5.404 ± 0.053 | 8.08 ± 0.084 |
3.853 ± 0.064 | 5.712 ± 0.064 |
1.159 ± 0.025 | 10.209 ± 0.085 |
10.556 ± 0.082 | 8.2 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.347 ± 0.034 | 7.534 ± 0.075 |
2.436 ± 0.035 | 2.922 ± 0.043 |
2.918 ± 0.048 | 5.808 ± 0.062 |
5.658 ± 0.102 | 6.016 ± 0.059 |
0.643 ± 0.019 | 4.503 ± 0.051 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
