
Methylobacterium sp. Leaf399
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Methylobacterium; unclassified Methylobacterium
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
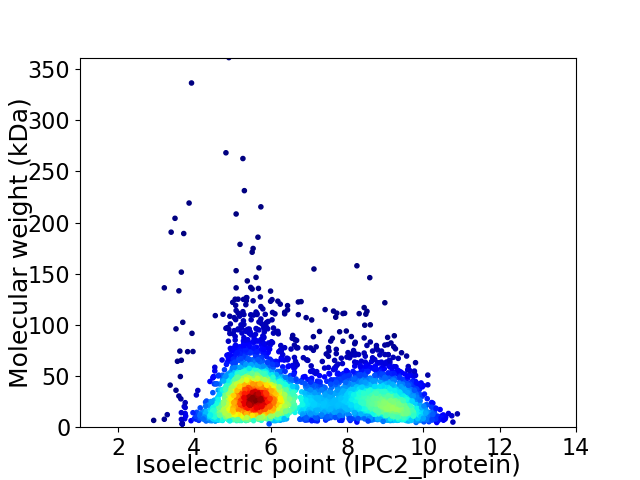
Virtual 2D-PAGE plot for 3885 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q5ZMA7|A0A0Q5ZMA7_9RHIZ Inositol monophosphatase OS=Methylobacterium sp. Leaf399 OX=1736364 GN=ASG40_18625 PE=4 SV=1
MM1 pKa = 6.91SQSVSNTQPSLVVKK15 pKa = 10.2SYY17 pKa = 9.16ITTQGLYY24 pKa = 7.73PTAGSGGPGTAPQNDD39 pKa = 4.24DD40 pKa = 3.39EE41 pKa = 5.05YY42 pKa = 10.59MGVIRR47 pKa = 11.84LTAASYY53 pKa = 11.32APGVDD58 pKa = 4.05VSGQTLNIRR67 pKa = 11.84GNEE70 pKa = 3.78ALYY73 pKa = 10.8SILGTRR79 pKa = 11.84YY80 pKa = 10.17GGDD83 pKa = 2.83GRR85 pKa = 11.84TTFKK89 pKa = 11.11LPDD92 pKa = 3.6FMGDD96 pKa = 3.09VSVGVGGGFLTGLGQHH112 pKa = 6.78GGTTATALTTANLPANLGGQGQTFDD137 pKa = 3.39NHH139 pKa = 5.95QSSEE143 pKa = 3.88AANYY147 pKa = 9.24LISVNGFFGTYY158 pKa = 9.49RR159 pKa = 11.84AGTLMQTAFDD169 pKa = 3.85IVPEE173 pKa = 4.43GFLPADD179 pKa = 3.4GRR181 pKa = 11.84LLNSADD187 pKa = 3.69YY188 pKa = 9.79PLLYY192 pKa = 10.4AALGTAYY199 pKa = 10.14GGDD202 pKa = 3.32GTTFRR207 pKa = 11.84LPDD210 pKa = 3.33LTGRR214 pKa = 11.84AVMGDD219 pKa = 3.59DD220 pKa = 3.39ASTVIGQQVGSATVSLTNANAPASAGGSGQAFANGQPALVMTYY263 pKa = 10.13LIAVQGLYY271 pKa = 9.9PDD273 pKa = 3.98RR274 pKa = 11.84DD275 pKa = 3.93GGRR278 pKa = 11.84GAYY281 pKa = 9.32SEE283 pKa = 4.66PYY285 pKa = 9.66IGEE288 pKa = 4.18LFAYY292 pKa = 10.13AGTDD296 pKa = 3.21VSDD299 pKa = 3.98ANLPEE304 pKa = 5.08GYY306 pKa = 10.15LRR308 pKa = 11.84AEE310 pKa = 4.39GQLLPINQYY319 pKa = 9.03QALYY323 pKa = 8.54TLIGATYY330 pKa = 8.95GGDD333 pKa = 3.65GISVFRR339 pKa = 11.84LPDD342 pKa = 3.08LRR344 pKa = 11.84GRR346 pKa = 11.84VAVDD350 pKa = 3.86DD351 pKa = 4.27GTDD354 pKa = 3.22RR355 pKa = 11.84PLGQTFGQAGEE366 pKa = 4.13TLDD369 pKa = 4.39ANEE372 pKa = 4.65IPAPAAPVITGPANGATTTDD392 pKa = 3.75DD393 pKa = 3.68TPDD396 pKa = 3.06ISGTASPNSIVTVVIDD412 pKa = 3.9GATVGTTRR420 pKa = 11.84ANASGAWTFTPTVKK434 pKa = 10.71LSDD437 pKa = 3.66AAHH440 pKa = 6.2SVTAAASYY448 pKa = 11.1GGDD451 pKa = 3.28QSPASTARR459 pKa = 11.84TFAVDD464 pKa = 3.74VNDD467 pKa = 4.21APALTGPQTALAAGSEE483 pKa = 4.17DD484 pKa = 3.13TAYY487 pKa = 9.79TVTAAQLLQGWTDD500 pKa = 3.4ADD502 pKa = 3.88FTTLAVTGLITSSGSVTSDD521 pKa = 2.81GNGTYY526 pKa = 9.79TVTPAANANGPVTLSYY542 pKa = 10.56QVSDD546 pKa = 3.48GVNAVSATRR555 pKa = 11.84TFTLAAINDD564 pKa = 3.99APQLTGPQATLPAGTEE580 pKa = 3.92DD581 pKa = 3.3TAFTVTSAQLLQGFSDD597 pKa = 4.09PEE599 pKa = 4.16GTTLVVSSLSVDD611 pKa = 3.04HH612 pKa = 6.24GTVVSDD618 pKa = 3.71GAGGYY623 pKa = 8.23TITPEE628 pKa = 3.81ANYY631 pKa = 10.24NGPLTISYY639 pKa = 10.61SVGDD643 pKa = 4.0GTASLPATLSTTLAAVNDD661 pKa = 4.02APRR664 pKa = 11.84LTGTQATLVVGTEE677 pKa = 3.96DD678 pKa = 3.12TAFTVTSAHH687 pKa = 6.79LLQGFSDD694 pKa = 4.31PDD696 pKa = 3.24GTTPVVLNLSVDD708 pKa = 3.24HH709 pKa = 6.17GTVVSDD715 pKa = 3.71GAGGYY720 pKa = 8.22TITPTANYY728 pKa = 9.21NGPLTISXSLSVDD741 pKa = 2.78HH742 pKa = 6.33GTVVSDD748 pKa = 3.71GAGGYY753 pKa = 8.22TITPTANYY761 pKa = 9.21NGPLTISYY769 pKa = 10.61SVGDD773 pKa = 4.11GTASTPATLSTAITAVNDD791 pKa = 3.67APRR794 pKa = 11.84LTDD797 pKa = 3.47TQATLADD804 pKa = 4.05GTEE807 pKa = 4.14DD808 pKa = 3.17TAFTVTSAQLLQGFSDD824 pKa = 3.91PDD826 pKa = 3.37GTTLVVSSLSVDD838 pKa = 3.04HH839 pKa = 6.24GTVVSDD845 pKa = 3.71GAGGYY850 pKa = 8.22TITPTANYY858 pKa = 9.21NGPLTISYY866 pKa = 10.61SVGDD870 pKa = 3.98GTASIPATLSTTLAAVNDD888 pKa = 3.97APRR891 pKa = 11.84LTGAQATLADD901 pKa = 4.13GTEE904 pKa = 4.2DD905 pKa = 3.25TTFTVTSAQLLQGFTDD921 pKa = 4.08PDD923 pKa = 3.33GTTPVLSNLSVDD935 pKa = 3.29HH936 pKa = 6.09GTVVSDD942 pKa = 3.71GAGGYY947 pKa = 8.22TITPTANYY955 pKa = 8.77NGPITISYY963 pKa = 10.49DD964 pKa = 3.24VGDD967 pKa = 4.31GVSSVAATRR976 pKa = 11.84DD977 pKa = 3.46VTIDD981 pKa = 3.48AVDD984 pKa = 4.06DD985 pKa = 4.47APVLAEE991 pKa = 4.74PGTVTLSEE999 pKa = 4.33SAAPGTTVATLSATDD1014 pKa = 3.76PDD1016 pKa = 4.3GAGVLTYY1023 pKa = 10.36AIQGGNQAGLFTIDD1037 pKa = 3.08ATTGVIALAAGRR1049 pKa = 11.84SVDD1052 pKa = 3.34AQTTARR1058 pKa = 11.84HH1059 pKa = 4.85VLGVRR1064 pKa = 11.84ATDD1067 pKa = 3.81GSGLVDD1073 pKa = 4.91DD1074 pKa = 5.47RR1075 pKa = 11.84SLTIDD1080 pKa = 3.42VLDD1083 pKa = 3.67VDD1085 pKa = 4.15RR1086 pKa = 11.84AGVLTLTGLTGGNARR1101 pKa = 11.84EE1102 pKa = 4.21GTPIVALLADD1112 pKa = 4.2ADD1114 pKa = 4.26EE1115 pKa = 4.59ATGAITYY1122 pKa = 7.51TFTDD1126 pKa = 3.26VSDD1129 pKa = 3.98PQAPRR1134 pKa = 11.84VLLTGSSASYY1144 pKa = 9.54TPDD1147 pKa = 2.86YY1148 pKa = 9.64TAAGQAIAVTVSYY1161 pKa = 10.94TDD1163 pKa = 3.82DD1164 pKa = 3.31QGHH1167 pKa = 6.44TGLASASAGVVGNVDD1182 pKa = 3.35VAPPQTQLVSGPAAVTRR1199 pKa = 11.84ATSATFTFSGSDD1211 pKa = 3.81DD1212 pKa = 3.58IAVDD1216 pKa = 3.7HH1217 pKa = 6.65YY1218 pKa = 11.06EE1219 pKa = 3.83VSLDD1223 pKa = 2.99GGGYY1227 pKa = 10.01VRR1229 pKa = 11.84TTSPYY1234 pKa = 9.59TVTGLADD1241 pKa = 3.93GVHH1244 pKa = 6.01SFAVRR1249 pKa = 11.84AVDD1252 pKa = 3.5AAGNADD1258 pKa = 4.32LTPEE1262 pKa = 4.15STLWTVDD1269 pKa = 3.18TQAPTVSGVSLSGPGIVRR1287 pKa = 11.84GSGTLTVGQSVIISVALSEE1306 pKa = 4.52PVTLTDD1312 pKa = 3.78AGGLRR1317 pKa = 11.84LQLSDD1322 pKa = 4.16GGVAAYY1328 pKa = 9.41DD1329 pKa = 3.68AAASDD1334 pKa = 3.77ASHH1337 pKa = 6.91LVFTHH1342 pKa = 5.49TVVVDD1347 pKa = 3.97AVSADD1352 pKa = 3.7LAITGLTLGSATILDD1367 pKa = 3.75AAGNVASLAGAATNPAGALRR1387 pKa = 11.84IDD1389 pKa = 5.09GYY1391 pKa = 9.62TGTAAAEE1398 pKa = 4.44TFHH1401 pKa = 6.07GTPGAEE1407 pKa = 4.06TFKK1410 pKa = 11.24GQGGNDD1416 pKa = 3.33TYY1418 pKa = 11.61LVDD1421 pKa = 3.72NVGDD1425 pKa = 3.8RR1426 pKa = 11.84VIEE1429 pKa = 4.07ATGGGSDD1436 pKa = 3.59SVIASVSYY1444 pKa = 10.62RR1445 pKa = 11.84LAAGQEE1451 pKa = 4.17IEE1453 pKa = 4.48SLALSTSTGGTALTLTGNEE1472 pKa = 4.15FANILTGNDD1481 pKa = 3.3GDD1483 pKa = 3.9NTLNGGLGADD1493 pKa = 3.91TMTGGAGSDD1502 pKa = 3.68FYY1504 pKa = 11.91VVDD1507 pKa = 4.14DD1508 pKa = 4.83AGDD1511 pKa = 3.52RR1512 pKa = 11.84AIEE1515 pKa = 3.95FAGLGTDD1522 pKa = 3.91LVYY1525 pKa = 10.41ATVSYY1530 pKa = 11.22SLGGSQLEE1538 pKa = 4.12NLTLAGTDD1546 pKa = 3.19NLTGLGNSLANVISGNGGDD1565 pKa = 3.73NVINGLGGADD1575 pKa = 3.47RR1576 pKa = 11.84MSGGAGSDD1584 pKa = 3.18TYY1586 pKa = 11.7YY1587 pKa = 11.18VDD1589 pKa = 4.09NLDD1592 pKa = 4.36DD1593 pKa = 4.41KK1594 pKa = 11.55VIEE1597 pKa = 4.08ATGAVGTDD1605 pKa = 3.05LVYY1608 pKa = 11.13ASVSYY1613 pKa = 11.04SLAGSHH1619 pKa = 6.99LEE1621 pKa = 3.82NLTLTGAGNLTATGNSLANVLTGNSGANVLDD1652 pKa = 3.87GLGGADD1658 pKa = 3.65TMSGGAGSDD1667 pKa = 3.21TYY1669 pKa = 11.74YY1670 pKa = 11.16VDD1672 pKa = 3.64TVDD1675 pKa = 5.8DD1676 pKa = 4.19RR1677 pKa = 11.84VLEE1680 pKa = 4.06ATGAVGTDD1688 pKa = 3.05LVYY1691 pKa = 11.13ASVSYY1696 pKa = 10.99SLAGAHH1702 pKa = 6.7LEE1704 pKa = 3.88NLTLTGAGNLTATGNSLANVITGTRR1729 pKa = 11.84GANILDD1735 pKa = 3.95GQGGDD1740 pKa = 4.86DD1741 pKa = 4.2ILTGGAGADD1750 pKa = 3.14TFVFKK1755 pKa = 9.98TALGSGNVDD1764 pKa = 4.04HH1765 pKa = 6.92ITDD1768 pKa = 4.72FVASTAAVHH1777 pKa = 6.48DD1778 pKa = 4.37TVQLARR1784 pKa = 11.84SVFATLAAGQLTEE1797 pKa = 5.1AAFKK1801 pKa = 10.96DD1802 pKa = 4.07SGNVDD1807 pKa = 4.25HH1808 pKa = 6.99ITDD1811 pKa = 4.72FVASTAAVHH1820 pKa = 6.48DD1821 pKa = 4.37TVQLARR1827 pKa = 11.84SVFATLAAGQLTEE1840 pKa = 5.3AAFKK1844 pKa = 11.0DD1845 pKa = 3.98LGVSAADD1852 pKa = 3.16ASDD1855 pKa = 4.31RR1856 pKa = 11.84ILYY1859 pKa = 10.33DD1860 pKa = 3.13KK1861 pKa = 10.68GTGGLFYY1868 pKa = 10.82DD1869 pKa = 4.32ADD1871 pKa = 4.28GSGSAATAVQFAVIDD1886 pKa = 3.88TKK1888 pKa = 10.82VTLSHH1893 pKa = 7.01HH1894 pKa = 6.78DD1895 pKa = 3.58FFVVV1899 pKa = 3.31
MM1 pKa = 6.91SQSVSNTQPSLVVKK15 pKa = 10.2SYY17 pKa = 9.16ITTQGLYY24 pKa = 7.73PTAGSGGPGTAPQNDD39 pKa = 4.24DD40 pKa = 3.39EE41 pKa = 5.05YY42 pKa = 10.59MGVIRR47 pKa = 11.84LTAASYY53 pKa = 11.32APGVDD58 pKa = 4.05VSGQTLNIRR67 pKa = 11.84GNEE70 pKa = 3.78ALYY73 pKa = 10.8SILGTRR79 pKa = 11.84YY80 pKa = 10.17GGDD83 pKa = 2.83GRR85 pKa = 11.84TTFKK89 pKa = 11.11LPDD92 pKa = 3.6FMGDD96 pKa = 3.09VSVGVGGGFLTGLGQHH112 pKa = 6.78GGTTATALTTANLPANLGGQGQTFDD137 pKa = 3.39NHH139 pKa = 5.95QSSEE143 pKa = 3.88AANYY147 pKa = 9.24LISVNGFFGTYY158 pKa = 9.49RR159 pKa = 11.84AGTLMQTAFDD169 pKa = 3.85IVPEE173 pKa = 4.43GFLPADD179 pKa = 3.4GRR181 pKa = 11.84LLNSADD187 pKa = 3.69YY188 pKa = 9.79PLLYY192 pKa = 10.4AALGTAYY199 pKa = 10.14GGDD202 pKa = 3.32GTTFRR207 pKa = 11.84LPDD210 pKa = 3.33LTGRR214 pKa = 11.84AVMGDD219 pKa = 3.59DD220 pKa = 3.39ASTVIGQQVGSATVSLTNANAPASAGGSGQAFANGQPALVMTYY263 pKa = 10.13LIAVQGLYY271 pKa = 9.9PDD273 pKa = 3.98RR274 pKa = 11.84DD275 pKa = 3.93GGRR278 pKa = 11.84GAYY281 pKa = 9.32SEE283 pKa = 4.66PYY285 pKa = 9.66IGEE288 pKa = 4.18LFAYY292 pKa = 10.13AGTDD296 pKa = 3.21VSDD299 pKa = 3.98ANLPEE304 pKa = 5.08GYY306 pKa = 10.15LRR308 pKa = 11.84AEE310 pKa = 4.39GQLLPINQYY319 pKa = 9.03QALYY323 pKa = 8.54TLIGATYY330 pKa = 8.95GGDD333 pKa = 3.65GISVFRR339 pKa = 11.84LPDD342 pKa = 3.08LRR344 pKa = 11.84GRR346 pKa = 11.84VAVDD350 pKa = 3.86DD351 pKa = 4.27GTDD354 pKa = 3.22RR355 pKa = 11.84PLGQTFGQAGEE366 pKa = 4.13TLDD369 pKa = 4.39ANEE372 pKa = 4.65IPAPAAPVITGPANGATTTDD392 pKa = 3.75DD393 pKa = 3.68TPDD396 pKa = 3.06ISGTASPNSIVTVVIDD412 pKa = 3.9GATVGTTRR420 pKa = 11.84ANASGAWTFTPTVKK434 pKa = 10.71LSDD437 pKa = 3.66AAHH440 pKa = 6.2SVTAAASYY448 pKa = 11.1GGDD451 pKa = 3.28QSPASTARR459 pKa = 11.84TFAVDD464 pKa = 3.74VNDD467 pKa = 4.21APALTGPQTALAAGSEE483 pKa = 4.17DD484 pKa = 3.13TAYY487 pKa = 9.79TVTAAQLLQGWTDD500 pKa = 3.4ADD502 pKa = 3.88FTTLAVTGLITSSGSVTSDD521 pKa = 2.81GNGTYY526 pKa = 9.79TVTPAANANGPVTLSYY542 pKa = 10.56QVSDD546 pKa = 3.48GVNAVSATRR555 pKa = 11.84TFTLAAINDD564 pKa = 3.99APQLTGPQATLPAGTEE580 pKa = 3.92DD581 pKa = 3.3TAFTVTSAQLLQGFSDD597 pKa = 4.09PEE599 pKa = 4.16GTTLVVSSLSVDD611 pKa = 3.04HH612 pKa = 6.24GTVVSDD618 pKa = 3.71GAGGYY623 pKa = 8.23TITPEE628 pKa = 3.81ANYY631 pKa = 10.24NGPLTISYY639 pKa = 10.61SVGDD643 pKa = 4.0GTASLPATLSTTLAAVNDD661 pKa = 4.02APRR664 pKa = 11.84LTGTQATLVVGTEE677 pKa = 3.96DD678 pKa = 3.12TAFTVTSAHH687 pKa = 6.79LLQGFSDD694 pKa = 4.31PDD696 pKa = 3.24GTTPVVLNLSVDD708 pKa = 3.24HH709 pKa = 6.17GTVVSDD715 pKa = 3.71GAGGYY720 pKa = 8.22TITPTANYY728 pKa = 9.21NGPLTISXSLSVDD741 pKa = 2.78HH742 pKa = 6.33GTVVSDD748 pKa = 3.71GAGGYY753 pKa = 8.22TITPTANYY761 pKa = 9.21NGPLTISYY769 pKa = 10.61SVGDD773 pKa = 4.11GTASTPATLSTAITAVNDD791 pKa = 3.67APRR794 pKa = 11.84LTDD797 pKa = 3.47TQATLADD804 pKa = 4.05GTEE807 pKa = 4.14DD808 pKa = 3.17TAFTVTSAQLLQGFSDD824 pKa = 3.91PDD826 pKa = 3.37GTTLVVSSLSVDD838 pKa = 3.04HH839 pKa = 6.24GTVVSDD845 pKa = 3.71GAGGYY850 pKa = 8.22TITPTANYY858 pKa = 9.21NGPLTISYY866 pKa = 10.61SVGDD870 pKa = 3.98GTASIPATLSTTLAAVNDD888 pKa = 3.97APRR891 pKa = 11.84LTGAQATLADD901 pKa = 4.13GTEE904 pKa = 4.2DD905 pKa = 3.25TTFTVTSAQLLQGFTDD921 pKa = 4.08PDD923 pKa = 3.33GTTPVLSNLSVDD935 pKa = 3.29HH936 pKa = 6.09GTVVSDD942 pKa = 3.71GAGGYY947 pKa = 8.22TITPTANYY955 pKa = 8.77NGPITISYY963 pKa = 10.49DD964 pKa = 3.24VGDD967 pKa = 4.31GVSSVAATRR976 pKa = 11.84DD977 pKa = 3.46VTIDD981 pKa = 3.48AVDD984 pKa = 4.06DD985 pKa = 4.47APVLAEE991 pKa = 4.74PGTVTLSEE999 pKa = 4.33SAAPGTTVATLSATDD1014 pKa = 3.76PDD1016 pKa = 4.3GAGVLTYY1023 pKa = 10.36AIQGGNQAGLFTIDD1037 pKa = 3.08ATTGVIALAAGRR1049 pKa = 11.84SVDD1052 pKa = 3.34AQTTARR1058 pKa = 11.84HH1059 pKa = 4.85VLGVRR1064 pKa = 11.84ATDD1067 pKa = 3.81GSGLVDD1073 pKa = 4.91DD1074 pKa = 5.47RR1075 pKa = 11.84SLTIDD1080 pKa = 3.42VLDD1083 pKa = 3.67VDD1085 pKa = 4.15RR1086 pKa = 11.84AGVLTLTGLTGGNARR1101 pKa = 11.84EE1102 pKa = 4.21GTPIVALLADD1112 pKa = 4.2ADD1114 pKa = 4.26EE1115 pKa = 4.59ATGAITYY1122 pKa = 7.51TFTDD1126 pKa = 3.26VSDD1129 pKa = 3.98PQAPRR1134 pKa = 11.84VLLTGSSASYY1144 pKa = 9.54TPDD1147 pKa = 2.86YY1148 pKa = 9.64TAAGQAIAVTVSYY1161 pKa = 10.94TDD1163 pKa = 3.82DD1164 pKa = 3.31QGHH1167 pKa = 6.44TGLASASAGVVGNVDD1182 pKa = 3.35VAPPQTQLVSGPAAVTRR1199 pKa = 11.84ATSATFTFSGSDD1211 pKa = 3.81DD1212 pKa = 3.58IAVDD1216 pKa = 3.7HH1217 pKa = 6.65YY1218 pKa = 11.06EE1219 pKa = 3.83VSLDD1223 pKa = 2.99GGGYY1227 pKa = 10.01VRR1229 pKa = 11.84TTSPYY1234 pKa = 9.59TVTGLADD1241 pKa = 3.93GVHH1244 pKa = 6.01SFAVRR1249 pKa = 11.84AVDD1252 pKa = 3.5AAGNADD1258 pKa = 4.32LTPEE1262 pKa = 4.15STLWTVDD1269 pKa = 3.18TQAPTVSGVSLSGPGIVRR1287 pKa = 11.84GSGTLTVGQSVIISVALSEE1306 pKa = 4.52PVTLTDD1312 pKa = 3.78AGGLRR1317 pKa = 11.84LQLSDD1322 pKa = 4.16GGVAAYY1328 pKa = 9.41DD1329 pKa = 3.68AAASDD1334 pKa = 3.77ASHH1337 pKa = 6.91LVFTHH1342 pKa = 5.49TVVVDD1347 pKa = 3.97AVSADD1352 pKa = 3.7LAITGLTLGSATILDD1367 pKa = 3.75AAGNVASLAGAATNPAGALRR1387 pKa = 11.84IDD1389 pKa = 5.09GYY1391 pKa = 9.62TGTAAAEE1398 pKa = 4.44TFHH1401 pKa = 6.07GTPGAEE1407 pKa = 4.06TFKK1410 pKa = 11.24GQGGNDD1416 pKa = 3.33TYY1418 pKa = 11.61LVDD1421 pKa = 3.72NVGDD1425 pKa = 3.8RR1426 pKa = 11.84VIEE1429 pKa = 4.07ATGGGSDD1436 pKa = 3.59SVIASVSYY1444 pKa = 10.62RR1445 pKa = 11.84LAAGQEE1451 pKa = 4.17IEE1453 pKa = 4.48SLALSTSTGGTALTLTGNEE1472 pKa = 4.15FANILTGNDD1481 pKa = 3.3GDD1483 pKa = 3.9NTLNGGLGADD1493 pKa = 3.91TMTGGAGSDD1502 pKa = 3.68FYY1504 pKa = 11.91VVDD1507 pKa = 4.14DD1508 pKa = 4.83AGDD1511 pKa = 3.52RR1512 pKa = 11.84AIEE1515 pKa = 3.95FAGLGTDD1522 pKa = 3.91LVYY1525 pKa = 10.41ATVSYY1530 pKa = 11.22SLGGSQLEE1538 pKa = 4.12NLTLAGTDD1546 pKa = 3.19NLTGLGNSLANVISGNGGDD1565 pKa = 3.73NVINGLGGADD1575 pKa = 3.47RR1576 pKa = 11.84MSGGAGSDD1584 pKa = 3.18TYY1586 pKa = 11.7YY1587 pKa = 11.18VDD1589 pKa = 4.09NLDD1592 pKa = 4.36DD1593 pKa = 4.41KK1594 pKa = 11.55VIEE1597 pKa = 4.08ATGAVGTDD1605 pKa = 3.05LVYY1608 pKa = 11.13ASVSYY1613 pKa = 11.04SLAGSHH1619 pKa = 6.99LEE1621 pKa = 3.82NLTLTGAGNLTATGNSLANVLTGNSGANVLDD1652 pKa = 3.87GLGGADD1658 pKa = 3.65TMSGGAGSDD1667 pKa = 3.21TYY1669 pKa = 11.74YY1670 pKa = 11.16VDD1672 pKa = 3.64TVDD1675 pKa = 5.8DD1676 pKa = 4.19RR1677 pKa = 11.84VLEE1680 pKa = 4.06ATGAVGTDD1688 pKa = 3.05LVYY1691 pKa = 11.13ASVSYY1696 pKa = 10.99SLAGAHH1702 pKa = 6.7LEE1704 pKa = 3.88NLTLTGAGNLTATGNSLANVITGTRR1729 pKa = 11.84GANILDD1735 pKa = 3.95GQGGDD1740 pKa = 4.86DD1741 pKa = 4.2ILTGGAGADD1750 pKa = 3.14TFVFKK1755 pKa = 9.98TALGSGNVDD1764 pKa = 4.04HH1765 pKa = 6.92ITDD1768 pKa = 4.72FVASTAAVHH1777 pKa = 6.48DD1778 pKa = 4.37TVQLARR1784 pKa = 11.84SVFATLAAGQLTEE1797 pKa = 5.1AAFKK1801 pKa = 10.96DD1802 pKa = 4.07SGNVDD1807 pKa = 4.25HH1808 pKa = 6.99ITDD1811 pKa = 4.72FVASTAAVHH1820 pKa = 6.48DD1821 pKa = 4.37TVQLARR1827 pKa = 11.84SVFATLAAGQLTEE1840 pKa = 5.3AAFKK1844 pKa = 11.0DD1845 pKa = 3.98LGVSAADD1852 pKa = 3.16ASDD1855 pKa = 4.31RR1856 pKa = 11.84ILYY1859 pKa = 10.33DD1860 pKa = 3.13KK1861 pKa = 10.68GTGGLFYY1868 pKa = 10.82DD1869 pKa = 4.32ADD1871 pKa = 4.28GSGSAATAVQFAVIDD1886 pKa = 3.88TKK1888 pKa = 10.82VTLSHH1893 pKa = 7.01HH1894 pKa = 6.78DD1895 pKa = 3.58FFVVV1899 pKa = 3.31
Molecular weight: 189.29 kDa
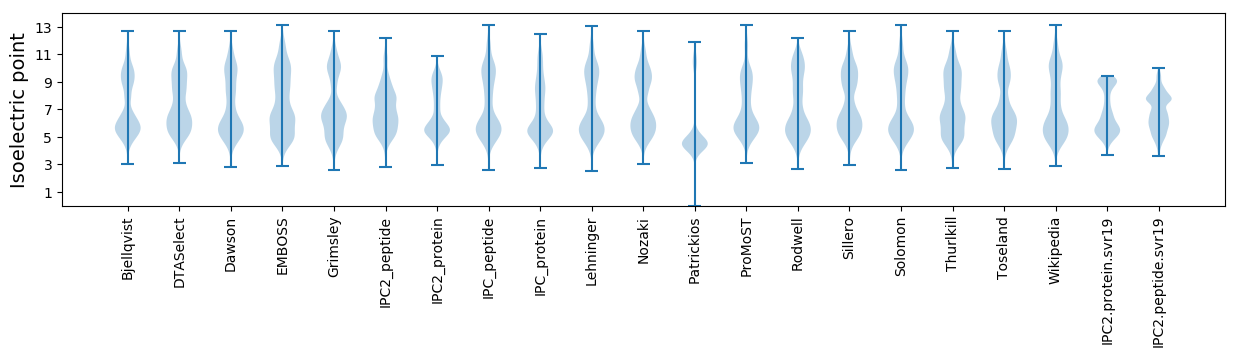
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q6A4Z3|A0A0Q6A4Z3_9RHIZ Large-conductance mechanosensitive channel OS=Methylobacterium sp. Leaf399 OX=1736364 GN=mscL PE=3 SV=1
MM1 pKa = 6.73TTPFSSRR8 pKa = 11.84RR9 pKa = 11.84NTLGLLALSGSLLVGFVALPQPAVARR35 pKa = 11.84NNTAVGVGTGAVAGALVAGPLGAVVGGIVGGVVGSNSEE73 pKa = 3.93SARR76 pKa = 11.84PRR78 pKa = 11.84RR79 pKa = 11.84SRR81 pKa = 11.84RR82 pKa = 11.84ARR84 pKa = 11.84AVRR87 pKa = 11.84PRR89 pKa = 11.84RR90 pKa = 11.84QPAVARR96 pKa = 11.84IAPRR100 pKa = 11.84AAPAARR106 pKa = 11.84PVAQAEE112 pKa = 4.12PRR114 pKa = 11.84PVSTVPTSTTWTNPRR129 pKa = 3.63
MM1 pKa = 6.73TTPFSSRR8 pKa = 11.84RR9 pKa = 11.84NTLGLLALSGSLLVGFVALPQPAVARR35 pKa = 11.84NNTAVGVGTGAVAGALVAGPLGAVVGGIVGGVVGSNSEE73 pKa = 3.93SARR76 pKa = 11.84PRR78 pKa = 11.84RR79 pKa = 11.84SRR81 pKa = 11.84RR82 pKa = 11.84ARR84 pKa = 11.84AVRR87 pKa = 11.84PRR89 pKa = 11.84RR90 pKa = 11.84QPAVARR96 pKa = 11.84IAPRR100 pKa = 11.84AAPAARR106 pKa = 11.84PVAQAEE112 pKa = 4.12PRR114 pKa = 11.84PVSTVPTSTTWTNPRR129 pKa = 3.63
Molecular weight: 13.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1232780 |
29 |
3564 |
317.3 |
33.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.17 ± 0.06 | 0.788 ± 0.013 |
5.91 ± 0.036 | 5.313 ± 0.04 |
3.321 ± 0.022 | 9.276 ± 0.05 |
1.889 ± 0.02 | 4.453 ± 0.028 |
2.532 ± 0.037 | 10.18 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.08 ± 0.018 | 2.115 ± 0.027 |
5.655 ± 0.043 | 2.678 ± 0.025 |
7.998 ± 0.048 | 4.981 ± 0.029 |
5.618 ± 0.039 | 7.897 ± 0.03 |
1.178 ± 0.017 | 1.968 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
