
Trametes coccinea BRFM310
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Polyporales; Polyporaceae; Trametes; Trametes coccinea
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
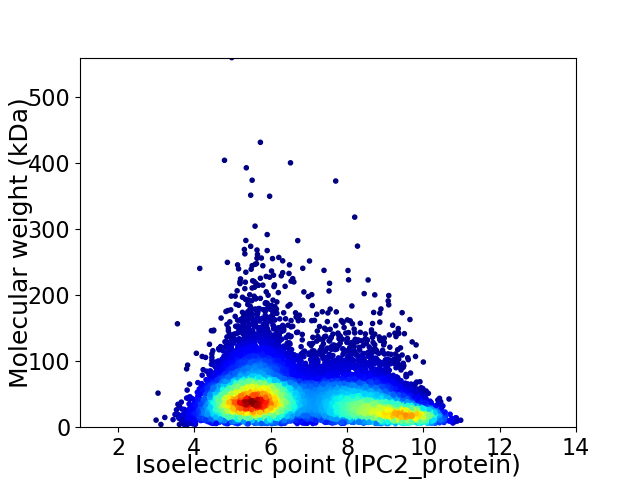
Virtual 2D-PAGE plot for 12648 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y2IP22|A0A1Y2IP22_PYCCO CRAL/TRIO domain-containing protein OS=Trametes coccinea BRFM310 OX=1353009 GN=PYCCODRAFT_1477715 PE=4 SV=1
MM1 pKa = 7.76CSLKK5 pKa = 10.6PSVSAALAASASATGVSGAASGSSSSSSSGNSSVGDD41 pKa = 3.39EE42 pKa = 4.44VVASTWYY49 pKa = 10.52AGWHH53 pKa = 6.25GEE55 pKa = 4.3DD56 pKa = 4.59FPPEE60 pKa = 4.21NISWSKK66 pKa = 8.18YY67 pKa = 6.24TQVTYY72 pKa = 11.12AFAVTTPDD80 pKa = 3.16VNTVSLEE87 pKa = 4.02ASDD90 pKa = 5.22EE91 pKa = 4.1EE92 pKa = 4.3LLPRR96 pKa = 11.84FVNAAHH102 pKa = 5.82QNNVKK107 pKa = 10.73AMLTVGGWTGSQYY120 pKa = 10.5FSSAVATEE128 pKa = 4.03ANRR131 pKa = 11.84TAFVKK136 pKa = 9.95TVLGLVSKK144 pKa = 11.08YY145 pKa = 10.9NLDD148 pKa = 3.58GLDD151 pKa = 4.4FDD153 pKa = 4.25WEE155 pKa = 4.5YY156 pKa = 10.9PNKK159 pKa = 10.16QGIGCNLISADD170 pKa = 3.91DD171 pKa = 3.89SQNFLSFLQALRR183 pKa = 11.84ADD185 pKa = 3.79PAGQNLTLSAAAGITPFAGPDD206 pKa = 3.73GTPMSDD212 pKa = 2.73VSDD215 pKa = 3.9FAKK218 pKa = 10.85VLDD221 pKa = 4.03YY222 pKa = 10.98VAIMNYY228 pKa = 9.64DD229 pKa = 2.97VWGSWSSSVGPNAPLNDD246 pKa = 3.78TCASAADD253 pKa = 3.86QQGSAVSAVKK263 pKa = 10.08AWTGAGFPANQLVLGVASYY282 pKa = 10.41GHH284 pKa = 6.2SFSVSNDD291 pKa = 2.84AALSGNTLLPYY302 pKa = 10.16PAFNKK307 pKa = 10.12ANQPLGDD314 pKa = 3.85KK315 pKa = 9.64WDD317 pKa = 3.64TDD319 pKa = 3.69AAPGVDD325 pKa = 3.15QCGNPTGGPSGIFDD339 pKa = 4.06FFGLIDD345 pKa = 4.34SKK347 pKa = 11.27FLTANGTVADD357 pKa = 4.96GIDD360 pKa = 3.45YY361 pKa = 10.92RR362 pKa = 11.84FDD364 pKa = 3.45EE365 pKa = 5.16CSQTPYY371 pKa = 10.61VYY373 pKa = 10.71NPSTQVMVSYY383 pKa = 11.44DD384 pKa = 3.68DD385 pKa = 3.74ATSFAAKK392 pKa = 10.22GKK394 pKa = 9.8FINDD398 pKa = 3.14AGLLGFAMWEE408 pKa = 3.97AAGDD412 pKa = 4.11SNDD415 pKa = 3.43ILLDD419 pKa = 4.56AIEE422 pKa = 3.89QAIGFEE428 pKa = 4.46EE429 pKa = 4.51VDD431 pKa = 3.79CC432 pKa = 5.43
MM1 pKa = 7.76CSLKK5 pKa = 10.6PSVSAALAASASATGVSGAASGSSSSSSSGNSSVGDD41 pKa = 3.39EE42 pKa = 4.44VVASTWYY49 pKa = 10.52AGWHH53 pKa = 6.25GEE55 pKa = 4.3DD56 pKa = 4.59FPPEE60 pKa = 4.21NISWSKK66 pKa = 8.18YY67 pKa = 6.24TQVTYY72 pKa = 11.12AFAVTTPDD80 pKa = 3.16VNTVSLEE87 pKa = 4.02ASDD90 pKa = 5.22EE91 pKa = 4.1EE92 pKa = 4.3LLPRR96 pKa = 11.84FVNAAHH102 pKa = 5.82QNNVKK107 pKa = 10.73AMLTVGGWTGSQYY120 pKa = 10.5FSSAVATEE128 pKa = 4.03ANRR131 pKa = 11.84TAFVKK136 pKa = 9.95TVLGLVSKK144 pKa = 11.08YY145 pKa = 10.9NLDD148 pKa = 3.58GLDD151 pKa = 4.4FDD153 pKa = 4.25WEE155 pKa = 4.5YY156 pKa = 10.9PNKK159 pKa = 10.16QGIGCNLISADD170 pKa = 3.91DD171 pKa = 3.89SQNFLSFLQALRR183 pKa = 11.84ADD185 pKa = 3.79PAGQNLTLSAAAGITPFAGPDD206 pKa = 3.73GTPMSDD212 pKa = 2.73VSDD215 pKa = 3.9FAKK218 pKa = 10.85VLDD221 pKa = 4.03YY222 pKa = 10.98VAIMNYY228 pKa = 9.64DD229 pKa = 2.97VWGSWSSSVGPNAPLNDD246 pKa = 3.78TCASAADD253 pKa = 3.86QQGSAVSAVKK263 pKa = 10.08AWTGAGFPANQLVLGVASYY282 pKa = 10.41GHH284 pKa = 6.2SFSVSNDD291 pKa = 2.84AALSGNTLLPYY302 pKa = 10.16PAFNKK307 pKa = 10.12ANQPLGDD314 pKa = 3.85KK315 pKa = 9.64WDD317 pKa = 3.64TDD319 pKa = 3.69AAPGVDD325 pKa = 3.15QCGNPTGGPSGIFDD339 pKa = 4.06FFGLIDD345 pKa = 4.34SKK347 pKa = 11.27FLTANGTVADD357 pKa = 4.96GIDD360 pKa = 3.45YY361 pKa = 10.92RR362 pKa = 11.84FDD364 pKa = 3.45EE365 pKa = 5.16CSQTPYY371 pKa = 10.61VYY373 pKa = 10.71NPSTQVMVSYY383 pKa = 11.44DD384 pKa = 3.68DD385 pKa = 3.74ATSFAAKK392 pKa = 10.22GKK394 pKa = 9.8FINDD398 pKa = 3.14AGLLGFAMWEE408 pKa = 3.97AAGDD412 pKa = 4.11SNDD415 pKa = 3.43ILLDD419 pKa = 4.56AIEE422 pKa = 3.89QAIGFEE428 pKa = 4.46EE429 pKa = 4.51VDD431 pKa = 3.79CC432 pKa = 5.43
Molecular weight: 45.06 kDa
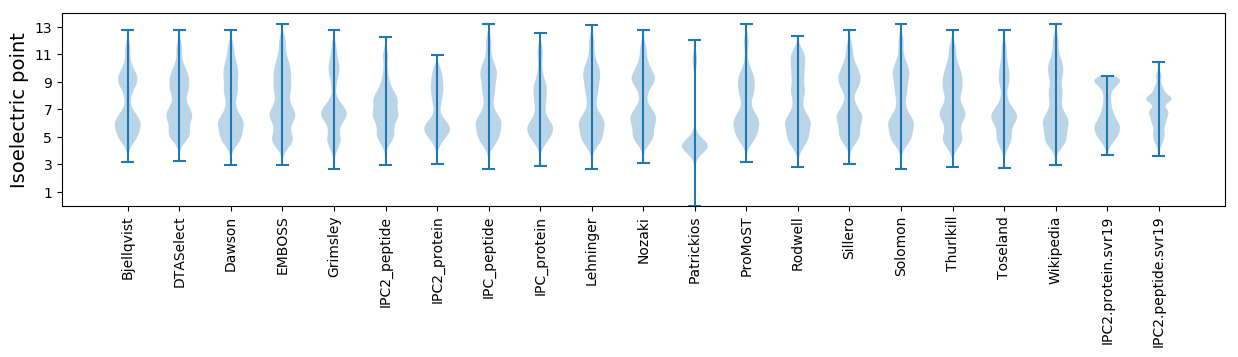
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y2IP82|A0A1Y2IP82_PYCCO Uncharacterized protein OS=Trametes coccinea BRFM310 OX=1353009 GN=PYCCODRAFT_1477236 PE=4 SV=1
MM1 pKa = 7.89PKK3 pKa = 9.88RR4 pKa = 11.84SARR7 pKa = 11.84KK8 pKa = 8.95RR9 pKa = 11.84RR10 pKa = 11.84LARR13 pKa = 11.84PSSRR17 pKa = 11.84SSSARR22 pKa = 11.84TTRR25 pKa = 11.84KK26 pKa = 9.15RR27 pKa = 11.84MKK29 pKa = 8.8RR30 pKa = 11.84TRR32 pKa = 11.84MTKK35 pKa = 10.27KK36 pKa = 8.4MWRR39 pKa = 11.84LLPSRR44 pKa = 11.84SRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84AVASEE53 pKa = 3.48RR54 pKa = 11.84HH55 pKa = 5.53RR56 pKa = 11.84PQSRR60 pKa = 11.84PEE62 pKa = 3.76RR63 pKa = 11.84RR64 pKa = 11.84WRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84WMRR71 pKa = 11.84TSTTSWRR78 pKa = 11.84TAMEE82 pKa = 4.32KK83 pKa = 9.88GTT85 pKa = 3.89
MM1 pKa = 7.89PKK3 pKa = 9.88RR4 pKa = 11.84SARR7 pKa = 11.84KK8 pKa = 8.95RR9 pKa = 11.84RR10 pKa = 11.84LARR13 pKa = 11.84PSSRR17 pKa = 11.84SSSARR22 pKa = 11.84TTRR25 pKa = 11.84KK26 pKa = 9.15RR27 pKa = 11.84MKK29 pKa = 8.8RR30 pKa = 11.84TRR32 pKa = 11.84MTKK35 pKa = 10.27KK36 pKa = 8.4MWRR39 pKa = 11.84LLPSRR44 pKa = 11.84SRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84AVASEE53 pKa = 3.48RR54 pKa = 11.84HH55 pKa = 5.53RR56 pKa = 11.84PQSRR60 pKa = 11.84PEE62 pKa = 3.76RR63 pKa = 11.84RR64 pKa = 11.84WRR66 pKa = 11.84RR67 pKa = 11.84RR68 pKa = 11.84WMRR71 pKa = 11.84TSTTSWRR78 pKa = 11.84TAMEE82 pKa = 4.32KK83 pKa = 9.88GTT85 pKa = 3.89
Molecular weight: 10.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5438953 |
39 |
5057 |
430.0 |
47.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.644 ± 0.022 | 1.284 ± 0.009 |
5.495 ± 0.014 | 5.886 ± 0.023 |
3.479 ± 0.014 | 6.564 ± 0.019 |
2.607 ± 0.011 | 4.344 ± 0.015 |
4.104 ± 0.022 | 9.103 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.034 ± 0.008 | 2.982 ± 0.012 |
7.089 ± 0.032 | 3.715 ± 0.016 |
6.825 ± 0.021 | 8.508 ± 0.036 |
5.853 ± 0.014 | 6.436 ± 0.017 |
1.431 ± 0.009 | 2.619 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
