
Coriobacteriaceae bacterium CHKCI002
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; unclassified Coriobacteriaceae
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
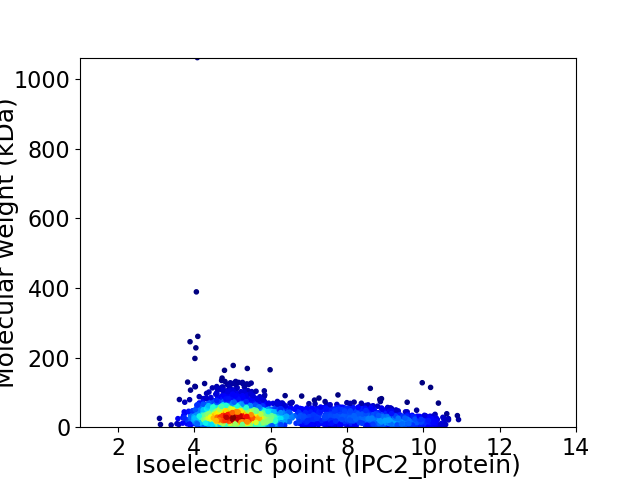
Virtual 2D-PAGE plot for 2836 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A143XB61|A0A143XB61_9ACTN Aspartokinase OS=Coriobacteriaceae bacterium CHKCI002 OX=1780377 GN=lysC PE=3 SV=1
MM1 pKa = 7.97RR2 pKa = 11.84DD3 pKa = 2.8SFAYY7 pKa = 10.2VGNEE11 pKa = 4.04TVGSMVEE18 pKa = 3.86ARR20 pKa = 11.84LGALGYY26 pKa = 10.47VRR28 pKa = 11.84ASDD31 pKa = 3.38VGSARR36 pKa = 11.84TVVTYY41 pKa = 7.55CTSQTALEE49 pKa = 4.23DD50 pKa = 3.89AYY52 pKa = 10.87FDD54 pKa = 3.98EE55 pKa = 5.77GGLVQAAAAGTLLVDD70 pKa = 5.43LSPSTPGFARR80 pKa = 11.84EE81 pKa = 4.07LNAVAAVSDD90 pKa = 4.17LVAVEE95 pKa = 4.48APLAVVDD102 pKa = 4.79PVRR105 pKa = 11.84ADD107 pKa = 3.25AFSDD111 pKa = 3.44KK112 pKa = 11.35GNLVCFAAGDD122 pKa = 3.92EE123 pKa = 4.46DD124 pKa = 4.19ALAAARR130 pKa = 11.84PVLEE134 pKa = 4.31AVAGSVRR141 pKa = 11.84DD142 pKa = 3.56MGGAGAGQLARR153 pKa = 11.84AAWTLQATAQIVSAVEE169 pKa = 3.49ADD171 pKa = 3.23ALYY174 pKa = 10.34RR175 pKa = 11.84AVRR178 pKa = 11.84RR179 pKa = 11.84SSSPFGEE186 pKa = 3.85EE187 pKa = 3.62ALRR190 pKa = 11.84VGAAAPLAEE199 pKa = 4.4SVLAAVDD206 pKa = 3.37EE207 pKa = 4.54GRR209 pKa = 11.84FDD211 pKa = 3.15GGYY214 pKa = 7.01TVEE217 pKa = 4.35MFMAEE222 pKa = 4.27LSAALTAADD231 pKa = 4.7DD232 pKa = 4.32ADD234 pKa = 5.37LILPQAEE241 pKa = 4.32ACLHH245 pKa = 5.78LLEE248 pKa = 4.91LLAVIGGSDD257 pKa = 3.54KK258 pKa = 11.34APSALALVYY267 pKa = 10.83GEE269 pKa = 4.55EE270 pKa = 4.18AACAEE275 pKa = 4.53QGLDD279 pKa = 3.05WTRR282 pKa = 11.84AEE284 pKa = 4.28SAFGEE289 pKa = 4.56GDD291 pKa = 4.25DD292 pKa = 4.75EE293 pKa = 4.76DD294 pKa = 6.61AEE296 pKa = 4.64DD297 pKa = 4.47GCEE300 pKa = 4.06EE301 pKa = 4.29GCDD304 pKa = 4.76ADD306 pKa = 4.68GCGCGHH312 pKa = 6.85AHH314 pKa = 6.76GGYY317 pKa = 9.16RR318 pKa = 11.84DD319 pKa = 3.59YY320 pKa = 11.14PDD322 pKa = 3.43YY323 pKa = 10.65PGYY326 pKa = 10.52GAYY329 pKa = 9.92SANN332 pKa = 3.47
MM1 pKa = 7.97RR2 pKa = 11.84DD3 pKa = 2.8SFAYY7 pKa = 10.2VGNEE11 pKa = 4.04TVGSMVEE18 pKa = 3.86ARR20 pKa = 11.84LGALGYY26 pKa = 10.47VRR28 pKa = 11.84ASDD31 pKa = 3.38VGSARR36 pKa = 11.84TVVTYY41 pKa = 7.55CTSQTALEE49 pKa = 4.23DD50 pKa = 3.89AYY52 pKa = 10.87FDD54 pKa = 3.98EE55 pKa = 5.77GGLVQAAAAGTLLVDD70 pKa = 5.43LSPSTPGFARR80 pKa = 11.84EE81 pKa = 4.07LNAVAAVSDD90 pKa = 4.17LVAVEE95 pKa = 4.48APLAVVDD102 pKa = 4.79PVRR105 pKa = 11.84ADD107 pKa = 3.25AFSDD111 pKa = 3.44KK112 pKa = 11.35GNLVCFAAGDD122 pKa = 3.92EE123 pKa = 4.46DD124 pKa = 4.19ALAAARR130 pKa = 11.84PVLEE134 pKa = 4.31AVAGSVRR141 pKa = 11.84DD142 pKa = 3.56MGGAGAGQLARR153 pKa = 11.84AAWTLQATAQIVSAVEE169 pKa = 3.49ADD171 pKa = 3.23ALYY174 pKa = 10.34RR175 pKa = 11.84AVRR178 pKa = 11.84RR179 pKa = 11.84SSSPFGEE186 pKa = 3.85EE187 pKa = 3.62ALRR190 pKa = 11.84VGAAAPLAEE199 pKa = 4.4SVLAAVDD206 pKa = 3.37EE207 pKa = 4.54GRR209 pKa = 11.84FDD211 pKa = 3.15GGYY214 pKa = 7.01TVEE217 pKa = 4.35MFMAEE222 pKa = 4.27LSAALTAADD231 pKa = 4.7DD232 pKa = 4.32ADD234 pKa = 5.37LILPQAEE241 pKa = 4.32ACLHH245 pKa = 5.78LLEE248 pKa = 4.91LLAVIGGSDD257 pKa = 3.54KK258 pKa = 11.34APSALALVYY267 pKa = 10.83GEE269 pKa = 4.55EE270 pKa = 4.18AACAEE275 pKa = 4.53QGLDD279 pKa = 3.05WTRR282 pKa = 11.84AEE284 pKa = 4.28SAFGEE289 pKa = 4.56GDD291 pKa = 4.25DD292 pKa = 4.75EE293 pKa = 4.76DD294 pKa = 6.61AEE296 pKa = 4.64DD297 pKa = 4.47GCEE300 pKa = 4.06EE301 pKa = 4.29GCDD304 pKa = 4.76ADD306 pKa = 4.68GCGCGHH312 pKa = 6.85AHH314 pKa = 6.76GGYY317 pKa = 9.16RR318 pKa = 11.84DD319 pKa = 3.59YY320 pKa = 11.14PDD322 pKa = 3.43YY323 pKa = 10.65PGYY326 pKa = 10.52GAYY329 pKa = 9.92SANN332 pKa = 3.47
Molecular weight: 33.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A143X0U1|A0A143X0U1_9ACTN Thioesterase superfamily protein OS=Coriobacteriaceae bacterium CHKCI002 OX=1780377 GN=BN3658_00646 PE=4 SV=1
MM1 pKa = 7.38TPASTAWTWWAPTRR15 pKa = 11.84TARR18 pKa = 11.84ARR20 pKa = 11.84LRR22 pKa = 11.84SRR24 pKa = 11.84SARR27 pKa = 11.84GRR29 pKa = 11.84RR30 pKa = 11.84SKK32 pKa = 9.64TATTTRR38 pKa = 11.84PPCCPASPCRR48 pKa = 11.84RR49 pKa = 11.84SSPGATGRR57 pKa = 11.84RR58 pKa = 11.84TTRR61 pKa = 11.84SRR63 pKa = 11.84SRR65 pKa = 11.84CTACAPWTRR74 pKa = 11.84RR75 pKa = 11.84ARR77 pKa = 11.84PLRR80 pKa = 11.84RR81 pKa = 11.84AHH83 pKa = 6.42RR84 pKa = 11.84CRR86 pKa = 11.84RR87 pKa = 11.84PARR90 pKa = 11.84FPSARR95 pKa = 11.84SMRR98 pKa = 11.84PRR100 pKa = 11.84KK101 pKa = 9.7RR102 pKa = 11.84PSPRR106 pKa = 11.84AAPWRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84SATSRR118 pKa = 11.84SPSRR122 pKa = 11.84APTSTACSSRR132 pKa = 11.84PRR134 pKa = 11.84TRR136 pKa = 11.84AATRR140 pKa = 11.84RR141 pKa = 11.84AASTTRR147 pKa = 11.84ARR149 pKa = 11.84STRR152 pKa = 11.84SQWPWPPTGRR162 pKa = 11.84ATSSPRR168 pKa = 11.84PRR170 pKa = 11.84LPLSPTTRR178 pKa = 11.84ASPPKK183 pKa = 10.28ARR185 pKa = 11.84AEE187 pKa = 4.21AGWPRR192 pKa = 11.84ASATPTRR199 pKa = 11.84IPRR202 pKa = 11.84SPPSTARR209 pKa = 11.84WTTPTRR215 pKa = 11.84AARR218 pKa = 11.84GLWKK222 pKa = 7.22TTCSRR227 pKa = 11.84SASKK231 pKa = 9.82PWATMPRR238 pKa = 11.84ARR240 pKa = 11.84RR241 pKa = 11.84APSSRR246 pKa = 11.84KK247 pKa = 9.06RR248 pKa = 11.84ATQAGCSRR256 pKa = 11.84SGRR259 pKa = 11.84PRR261 pKa = 11.84SRR263 pKa = 11.84TTTIWRR269 pKa = 11.84ARR271 pKa = 11.84RR272 pKa = 11.84SPTRR276 pKa = 11.84CGRR279 pKa = 11.84TSTRR283 pKa = 11.84ALATARR289 pKa = 11.84TEE291 pKa = 4.1PRR293 pKa = 11.84PHH295 pKa = 6.0LTGG298 pKa = 3.55
MM1 pKa = 7.38TPASTAWTWWAPTRR15 pKa = 11.84TARR18 pKa = 11.84ARR20 pKa = 11.84LRR22 pKa = 11.84SRR24 pKa = 11.84SARR27 pKa = 11.84GRR29 pKa = 11.84RR30 pKa = 11.84SKK32 pKa = 9.64TATTTRR38 pKa = 11.84PPCCPASPCRR48 pKa = 11.84RR49 pKa = 11.84SSPGATGRR57 pKa = 11.84RR58 pKa = 11.84TTRR61 pKa = 11.84SRR63 pKa = 11.84SRR65 pKa = 11.84CTACAPWTRR74 pKa = 11.84RR75 pKa = 11.84ARR77 pKa = 11.84PLRR80 pKa = 11.84RR81 pKa = 11.84AHH83 pKa = 6.42RR84 pKa = 11.84CRR86 pKa = 11.84RR87 pKa = 11.84PARR90 pKa = 11.84FPSARR95 pKa = 11.84SMRR98 pKa = 11.84PRR100 pKa = 11.84KK101 pKa = 9.7RR102 pKa = 11.84PSPRR106 pKa = 11.84AAPWRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84SATSRR118 pKa = 11.84SPSRR122 pKa = 11.84APTSTACSSRR132 pKa = 11.84PRR134 pKa = 11.84TRR136 pKa = 11.84AATRR140 pKa = 11.84RR141 pKa = 11.84AASTTRR147 pKa = 11.84ARR149 pKa = 11.84STRR152 pKa = 11.84SQWPWPPTGRR162 pKa = 11.84ATSSPRR168 pKa = 11.84PRR170 pKa = 11.84LPLSPTTRR178 pKa = 11.84ASPPKK183 pKa = 10.28ARR185 pKa = 11.84AEE187 pKa = 4.21AGWPRR192 pKa = 11.84ASATPTRR199 pKa = 11.84IPRR202 pKa = 11.84SPPSTARR209 pKa = 11.84WTTPTRR215 pKa = 11.84AARR218 pKa = 11.84GLWKK222 pKa = 7.22TTCSRR227 pKa = 11.84SASKK231 pKa = 9.82PWATMPRR238 pKa = 11.84ARR240 pKa = 11.84RR241 pKa = 11.84APSSRR246 pKa = 11.84KK247 pKa = 9.06RR248 pKa = 11.84ATQAGCSRR256 pKa = 11.84SGRR259 pKa = 11.84PRR261 pKa = 11.84SRR263 pKa = 11.84TTTIWRR269 pKa = 11.84ARR271 pKa = 11.84RR272 pKa = 11.84SPTRR276 pKa = 11.84CGRR279 pKa = 11.84TSTRR283 pKa = 11.84ALATARR289 pKa = 11.84TEE291 pKa = 4.1PRR293 pKa = 11.84PHH295 pKa = 6.0LTGG298 pKa = 3.55
Molecular weight: 33.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
947897 |
29 |
9826 |
334.2 |
36.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.807 ± 0.089 | 1.653 ± 0.025 |
5.955 ± 0.053 | 6.675 ± 0.046 |
3.828 ± 0.031 | 8.597 ± 0.049 |
1.721 ± 0.022 | 4.301 ± 0.042 |
3.43 ± 0.041 | 9.26 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.535 ± 0.024 | 2.583 ± 0.03 |
4.464 ± 0.031 | 2.636 ± 0.027 |
6.725 ± 0.068 | 5.838 ± 0.043 |
4.901 ± 0.045 | 8.224 ± 0.049 |
1.152 ± 0.019 | 2.715 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
