
Discula destructiva virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Gammapartitivirus
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
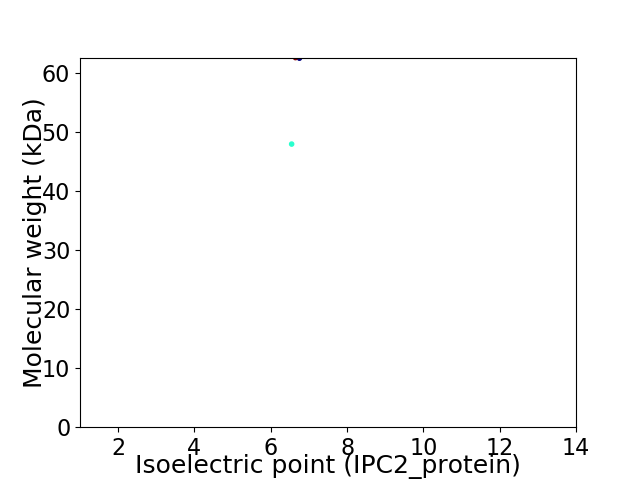
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q91HI8|Q91HI8_9VIRU RNA-dependent RNA polymerase OS=Discula destructiva virus 2 OX=160484 PE=4 SV=1
MM1 pKa = 7.69EE2 pKa = 6.06GFTQEE7 pKa = 4.09PTNTTVLAEE16 pKa = 4.2EE17 pKa = 4.25LHH19 pKa = 7.05SVDD22 pKa = 4.03TLHH25 pKa = 7.0LRR27 pKa = 11.84PGKK30 pKa = 8.56TRR32 pKa = 11.84STTSEE37 pKa = 3.76DD38 pKa = 3.45VLPNNYY44 pKa = 9.04EE45 pKa = 4.07DD46 pKa = 3.66PCLRR50 pKa = 11.84EE51 pKa = 3.35IAKK54 pKa = 10.49YY55 pKa = 10.25GGYY58 pKa = 7.94STYY61 pKa = 11.11SSNSNTDD68 pKa = 2.65PWIRR72 pKa = 11.84EE73 pKa = 3.95TLKK76 pKa = 10.91LHH78 pKa = 7.03DD79 pKa = 3.79RR80 pKa = 11.84QIYY83 pKa = 8.79EE84 pKa = 4.81DD85 pKa = 2.92IWGKK89 pKa = 6.99TRR91 pKa = 11.84RR92 pKa = 11.84PEE94 pKa = 4.14GTPGMYY100 pKa = 9.9KK101 pKa = 10.46ALGRR105 pKa = 11.84FGGEE109 pKa = 3.08RR110 pKa = 11.84CGFDD114 pKa = 5.05DD115 pKa = 6.4LSSQQKK121 pKa = 9.74SSMRR125 pKa = 11.84RR126 pKa = 11.84AIAKK130 pKa = 9.38AKK132 pKa = 10.28KK133 pKa = 9.98AFKK136 pKa = 10.34LPYY139 pKa = 9.55KK140 pKa = 10.34RR141 pKa = 11.84EE142 pKa = 3.91PLDD145 pKa = 3.15WHH147 pKa = 6.48EE148 pKa = 4.21VGQFLRR154 pKa = 11.84RR155 pKa = 11.84DD156 pKa = 3.54TSAGSTFMGSKK167 pKa = 10.02KK168 pKa = 10.11GDD170 pKa = 3.51VMEE173 pKa = 5.35EE174 pKa = 4.02IYY176 pKa = 10.94SEE178 pKa = 4.21ARR180 pKa = 11.84WLGHH184 pKa = 6.32RR185 pKa = 11.84MKK187 pKa = 10.64QDD189 pKa = 2.55GRR191 pKa = 11.84SRR193 pKa = 11.84FDD195 pKa = 3.2PTKK198 pKa = 10.31MRR200 pKa = 11.84FPPCLAGQRR209 pKa = 11.84GGMSDD214 pKa = 3.86RR215 pKa = 11.84DD216 pKa = 3.83DD217 pKa = 3.97PKK219 pKa = 11.11TRR221 pKa = 11.84LIWIYY226 pKa = 9.13PAEE229 pKa = 4.11MLCVEE234 pKa = 5.06GFYY237 pKa = 11.1APLMYY242 pKa = 9.21RR243 pKa = 11.84DD244 pKa = 4.18YY245 pKa = 10.85MSDD248 pKa = 3.43PNSPMLNGKK257 pKa = 9.37SSQRR261 pKa = 11.84LYY263 pKa = 10.77TEE265 pKa = 4.24WCCNLRR271 pKa = 11.84EE272 pKa = 4.15GEE274 pKa = 4.27TLYY277 pKa = 11.45GIDD280 pKa = 4.95FSAFDD285 pKa = 3.86SKK287 pKa = 11.33VPAWLIRR294 pKa = 11.84TAFDD298 pKa = 3.79IVKK301 pKa = 10.36QNINFEE307 pKa = 4.33TFEE310 pKa = 4.31GKK312 pKa = 9.17PVNKK316 pKa = 9.76VDD318 pKa = 3.27AQKK321 pKa = 10.5WKK323 pKa = 10.38NVWDD327 pKa = 3.61AMVWYY332 pKa = 8.66FINTPILMPDD342 pKa = 2.41GRR344 pKa = 11.84MFRR347 pKa = 11.84KK348 pKa = 9.98YY349 pKa = 10.14RR350 pKa = 11.84GVPSGSWWTQIIDD363 pKa = 3.75SVVNNILIDD372 pKa = 3.85YY373 pKa = 9.9LADD376 pKa = 3.91CQSVKK381 pKa = 10.15IPKK384 pKa = 9.84PEE386 pKa = 3.83VLGDD390 pKa = 3.64DD391 pKa = 3.6SAFRR395 pKa = 11.84SNDD398 pKa = 3.32QFDD401 pKa = 4.65LEE403 pKa = 4.49VAKK406 pKa = 10.44DD407 pKa = 3.64DD408 pKa = 4.05CVPTGMVIRR417 pKa = 11.84PEE419 pKa = 4.03KK420 pKa = 10.4CEE422 pKa = 3.91KK423 pKa = 10.45TEE425 pKa = 4.52DD426 pKa = 3.63PAEE429 pKa = 4.22FKK431 pKa = 11.21LLGTKK436 pKa = 9.45YY437 pKa = 10.21RR438 pKa = 11.84SGRR441 pKa = 11.84VHH443 pKa = 7.52RR444 pKa = 11.84STDD447 pKa = 2.79EE448 pKa = 3.96WFSLALYY455 pKa = 9.44PEE457 pKa = 4.56SSVLSLDD464 pKa = 3.43VSFTRR469 pKa = 11.84LVGLWLGGAMWDD481 pKa = 3.56KK482 pKa = 10.66QFCEE486 pKa = 4.26FMDD489 pKa = 4.49YY490 pKa = 10.98YY491 pKa = 9.16QTSYY495 pKa = 10.78PVPEE499 pKa = 4.77EE500 pKa = 3.98GWFSKK505 pKa = 10.13DD506 pKa = 3.02QKK508 pKa = 10.51RR509 pKa = 11.84WLEE512 pKa = 3.89IIYY515 pKa = 10.23SGKK518 pKa = 10.28APRR521 pKa = 11.84GWTTKK526 pKa = 10.52KK527 pKa = 10.35SLFWRR532 pKa = 11.84SIFYY536 pKa = 10.51AYY538 pKa = 10.44GG539 pKa = 3.3
MM1 pKa = 7.69EE2 pKa = 6.06GFTQEE7 pKa = 4.09PTNTTVLAEE16 pKa = 4.2EE17 pKa = 4.25LHH19 pKa = 7.05SVDD22 pKa = 4.03TLHH25 pKa = 7.0LRR27 pKa = 11.84PGKK30 pKa = 8.56TRR32 pKa = 11.84STTSEE37 pKa = 3.76DD38 pKa = 3.45VLPNNYY44 pKa = 9.04EE45 pKa = 4.07DD46 pKa = 3.66PCLRR50 pKa = 11.84EE51 pKa = 3.35IAKK54 pKa = 10.49YY55 pKa = 10.25GGYY58 pKa = 7.94STYY61 pKa = 11.11SSNSNTDD68 pKa = 2.65PWIRR72 pKa = 11.84EE73 pKa = 3.95TLKK76 pKa = 10.91LHH78 pKa = 7.03DD79 pKa = 3.79RR80 pKa = 11.84QIYY83 pKa = 8.79EE84 pKa = 4.81DD85 pKa = 2.92IWGKK89 pKa = 6.99TRR91 pKa = 11.84RR92 pKa = 11.84PEE94 pKa = 4.14GTPGMYY100 pKa = 9.9KK101 pKa = 10.46ALGRR105 pKa = 11.84FGGEE109 pKa = 3.08RR110 pKa = 11.84CGFDD114 pKa = 5.05DD115 pKa = 6.4LSSQQKK121 pKa = 9.74SSMRR125 pKa = 11.84RR126 pKa = 11.84AIAKK130 pKa = 9.38AKK132 pKa = 10.28KK133 pKa = 9.98AFKK136 pKa = 10.34LPYY139 pKa = 9.55KK140 pKa = 10.34RR141 pKa = 11.84EE142 pKa = 3.91PLDD145 pKa = 3.15WHH147 pKa = 6.48EE148 pKa = 4.21VGQFLRR154 pKa = 11.84RR155 pKa = 11.84DD156 pKa = 3.54TSAGSTFMGSKK167 pKa = 10.02KK168 pKa = 10.11GDD170 pKa = 3.51VMEE173 pKa = 5.35EE174 pKa = 4.02IYY176 pKa = 10.94SEE178 pKa = 4.21ARR180 pKa = 11.84WLGHH184 pKa = 6.32RR185 pKa = 11.84MKK187 pKa = 10.64QDD189 pKa = 2.55GRR191 pKa = 11.84SRR193 pKa = 11.84FDD195 pKa = 3.2PTKK198 pKa = 10.31MRR200 pKa = 11.84FPPCLAGQRR209 pKa = 11.84GGMSDD214 pKa = 3.86RR215 pKa = 11.84DD216 pKa = 3.83DD217 pKa = 3.97PKK219 pKa = 11.11TRR221 pKa = 11.84LIWIYY226 pKa = 9.13PAEE229 pKa = 4.11MLCVEE234 pKa = 5.06GFYY237 pKa = 11.1APLMYY242 pKa = 9.21RR243 pKa = 11.84DD244 pKa = 4.18YY245 pKa = 10.85MSDD248 pKa = 3.43PNSPMLNGKK257 pKa = 9.37SSQRR261 pKa = 11.84LYY263 pKa = 10.77TEE265 pKa = 4.24WCCNLRR271 pKa = 11.84EE272 pKa = 4.15GEE274 pKa = 4.27TLYY277 pKa = 11.45GIDD280 pKa = 4.95FSAFDD285 pKa = 3.86SKK287 pKa = 11.33VPAWLIRR294 pKa = 11.84TAFDD298 pKa = 3.79IVKK301 pKa = 10.36QNINFEE307 pKa = 4.33TFEE310 pKa = 4.31GKK312 pKa = 9.17PVNKK316 pKa = 9.76VDD318 pKa = 3.27AQKK321 pKa = 10.5WKK323 pKa = 10.38NVWDD327 pKa = 3.61AMVWYY332 pKa = 8.66FINTPILMPDD342 pKa = 2.41GRR344 pKa = 11.84MFRR347 pKa = 11.84KK348 pKa = 9.98YY349 pKa = 10.14RR350 pKa = 11.84GVPSGSWWTQIIDD363 pKa = 3.75SVVNNILIDD372 pKa = 3.85YY373 pKa = 9.9LADD376 pKa = 3.91CQSVKK381 pKa = 10.15IPKK384 pKa = 9.84PEE386 pKa = 3.83VLGDD390 pKa = 3.64DD391 pKa = 3.6SAFRR395 pKa = 11.84SNDD398 pKa = 3.32QFDD401 pKa = 4.65LEE403 pKa = 4.49VAKK406 pKa = 10.44DD407 pKa = 3.64DD408 pKa = 4.05CVPTGMVIRR417 pKa = 11.84PEE419 pKa = 4.03KK420 pKa = 10.4CEE422 pKa = 3.91KK423 pKa = 10.45TEE425 pKa = 4.52DD426 pKa = 3.63PAEE429 pKa = 4.22FKK431 pKa = 11.21LLGTKK436 pKa = 9.45YY437 pKa = 10.21RR438 pKa = 11.84SGRR441 pKa = 11.84VHH443 pKa = 7.52RR444 pKa = 11.84STDD447 pKa = 2.79EE448 pKa = 3.96WFSLALYY455 pKa = 9.44PEE457 pKa = 4.56SSVLSLDD464 pKa = 3.43VSFTRR469 pKa = 11.84LVGLWLGGAMWDD481 pKa = 3.56KK482 pKa = 10.66QFCEE486 pKa = 4.26FMDD489 pKa = 4.49YY490 pKa = 10.98YY491 pKa = 9.16QTSYY495 pKa = 10.78PVPEE499 pKa = 4.77EE500 pKa = 3.98GWFSKK505 pKa = 10.13DD506 pKa = 3.02QKK508 pKa = 10.51RR509 pKa = 11.84WLEE512 pKa = 3.89IIYY515 pKa = 10.23SGKK518 pKa = 10.28APRR521 pKa = 11.84GWTTKK526 pKa = 10.52KK527 pKa = 10.35SLFWRR532 pKa = 11.84SIFYY536 pKa = 10.51AYY538 pKa = 10.44GG539 pKa = 3.3
Molecular weight: 62.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q91HI8|Q91HI8_9VIRU RNA-dependent RNA polymerase OS=Discula destructiva virus 2 OX=160484 PE=4 SV=1
MM1 pKa = 8.05ADD3 pKa = 3.19TSSNIPSSVTPNDD16 pKa = 3.55SVSNSGKK23 pKa = 10.13RR24 pKa = 11.84KK25 pKa = 9.1SKK27 pKa = 9.65PGKK30 pKa = 9.85AEE32 pKa = 3.59RR33 pKa = 11.84LARR36 pKa = 11.84RR37 pKa = 11.84SAVGSQPGQAASASKK52 pKa = 10.74AAMFSSGSMAPKK64 pKa = 9.01PQPGKK69 pKa = 10.6YY70 pKa = 9.01PVVFQTGAGEE80 pKa = 4.06PSRR83 pKa = 11.84DD84 pKa = 3.42QQFAISGPTISKK96 pKa = 8.61TLEE99 pKa = 4.01GFPEE103 pKa = 4.03RR104 pKa = 11.84FSFSEE109 pKa = 3.99KK110 pKa = 9.22FTEE113 pKa = 5.35FKK115 pKa = 11.03ANSGFDD121 pKa = 4.04DD122 pKa = 4.26EE123 pKa = 6.08DD124 pKa = 4.43FEE126 pKa = 6.18LDD128 pKa = 3.38IVVSTLLRR136 pKa = 11.84LSQQVVHH143 pKa = 5.79SHH145 pKa = 5.44VNMGLPQGDD154 pKa = 4.3FAPVASTEE162 pKa = 3.82VRR164 pKa = 11.84VPGSVAAFVEE174 pKa = 4.44QFGEE178 pKa = 4.17HH179 pKa = 5.91SVPAIGTRR187 pKa = 11.84FLFKK191 pKa = 10.39DD192 pKa = 3.28YY193 pKa = 10.4RR194 pKa = 11.84QTVSRR199 pKa = 11.84LVWAAEE205 pKa = 3.69QAAVGKK211 pKa = 9.22WKK213 pKa = 10.65EE214 pKa = 3.7PLEE217 pKa = 4.16RR218 pKa = 11.84LWLPMSSSDD227 pKa = 3.05GHH229 pKa = 5.91TKK231 pKa = 10.86LEE233 pKa = 3.9IAHH236 pKa = 6.97RR237 pKa = 11.84LNAFLEE243 pKa = 4.29NAEE246 pKa = 4.02VQIPYY251 pKa = 10.34SILEE255 pKa = 4.54DD256 pKa = 3.78GVLSGTVPDD265 pKa = 3.59AWNGIKK271 pKa = 10.55GFLGDD276 pKa = 4.14PPTPGGVDD284 pKa = 2.75RR285 pKa = 11.84RR286 pKa = 11.84DD287 pKa = 3.29RR288 pKa = 11.84FDD290 pKa = 5.28FLFKK294 pKa = 10.57SYY296 pKa = 11.41NDD298 pKa = 3.7APQLAVAFTTAAATAVLGEE317 pKa = 4.61LRR319 pKa = 11.84LPWSAPSAGHH329 pKa = 6.73LNWSFNARR337 pKa = 11.84EE338 pKa = 4.01AFTRR342 pKa = 11.84LSDD345 pKa = 2.8NWARR349 pKa = 11.84KK350 pKa = 6.88STTYY354 pKa = 11.35AKK356 pKa = 10.34FFEE359 pKa = 4.91LSSSLSNRR367 pKa = 11.84TAATGSQSQMALVSEE382 pKa = 4.46SDD384 pKa = 3.77GITVVKK390 pKa = 9.07TALALSPPEE399 pKa = 4.09FSLVACFPASCLYY412 pKa = 10.66SGEE415 pKa = 3.96LVRR418 pKa = 11.84RR419 pKa = 11.84VVVTTPLSVRR429 pKa = 11.84QRR431 pKa = 11.84ATEE434 pKa = 4.17FVQMDD439 pKa = 3.24WRR441 pKa = 11.84AA442 pKa = 2.9
MM1 pKa = 8.05ADD3 pKa = 3.19TSSNIPSSVTPNDD16 pKa = 3.55SVSNSGKK23 pKa = 10.13RR24 pKa = 11.84KK25 pKa = 9.1SKK27 pKa = 9.65PGKK30 pKa = 9.85AEE32 pKa = 3.59RR33 pKa = 11.84LARR36 pKa = 11.84RR37 pKa = 11.84SAVGSQPGQAASASKK52 pKa = 10.74AAMFSSGSMAPKK64 pKa = 9.01PQPGKK69 pKa = 10.6YY70 pKa = 9.01PVVFQTGAGEE80 pKa = 4.06PSRR83 pKa = 11.84DD84 pKa = 3.42QQFAISGPTISKK96 pKa = 8.61TLEE99 pKa = 4.01GFPEE103 pKa = 4.03RR104 pKa = 11.84FSFSEE109 pKa = 3.99KK110 pKa = 9.22FTEE113 pKa = 5.35FKK115 pKa = 11.03ANSGFDD121 pKa = 4.04DD122 pKa = 4.26EE123 pKa = 6.08DD124 pKa = 4.43FEE126 pKa = 6.18LDD128 pKa = 3.38IVVSTLLRR136 pKa = 11.84LSQQVVHH143 pKa = 5.79SHH145 pKa = 5.44VNMGLPQGDD154 pKa = 4.3FAPVASTEE162 pKa = 3.82VRR164 pKa = 11.84VPGSVAAFVEE174 pKa = 4.44QFGEE178 pKa = 4.17HH179 pKa = 5.91SVPAIGTRR187 pKa = 11.84FLFKK191 pKa = 10.39DD192 pKa = 3.28YY193 pKa = 10.4RR194 pKa = 11.84QTVSRR199 pKa = 11.84LVWAAEE205 pKa = 3.69QAAVGKK211 pKa = 9.22WKK213 pKa = 10.65EE214 pKa = 3.7PLEE217 pKa = 4.16RR218 pKa = 11.84LWLPMSSSDD227 pKa = 3.05GHH229 pKa = 5.91TKK231 pKa = 10.86LEE233 pKa = 3.9IAHH236 pKa = 6.97RR237 pKa = 11.84LNAFLEE243 pKa = 4.29NAEE246 pKa = 4.02VQIPYY251 pKa = 10.34SILEE255 pKa = 4.54DD256 pKa = 3.78GVLSGTVPDD265 pKa = 3.59AWNGIKK271 pKa = 10.55GFLGDD276 pKa = 4.14PPTPGGVDD284 pKa = 2.75RR285 pKa = 11.84RR286 pKa = 11.84DD287 pKa = 3.29RR288 pKa = 11.84FDD290 pKa = 5.28FLFKK294 pKa = 10.57SYY296 pKa = 11.41NDD298 pKa = 3.7APQLAVAFTTAAATAVLGEE317 pKa = 4.61LRR319 pKa = 11.84LPWSAPSAGHH329 pKa = 6.73LNWSFNARR337 pKa = 11.84EE338 pKa = 4.01AFTRR342 pKa = 11.84LSDD345 pKa = 2.8NWARR349 pKa = 11.84KK350 pKa = 6.88STTYY354 pKa = 11.35AKK356 pKa = 10.34FFEE359 pKa = 4.91LSSSLSNRR367 pKa = 11.84TAATGSQSQMALVSEE382 pKa = 4.46SDD384 pKa = 3.77GITVVKK390 pKa = 9.07TALALSPPEE399 pKa = 4.09FSLVACFPASCLYY412 pKa = 10.66SGEE415 pKa = 3.96LVRR418 pKa = 11.84RR419 pKa = 11.84VVVTTPLSVRR429 pKa = 11.84QRR431 pKa = 11.84ATEE434 pKa = 4.17FVQMDD439 pKa = 3.24WRR441 pKa = 11.84AA442 pKa = 2.9
Molecular weight: 47.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
981 |
442 |
539 |
490.5 |
55.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.441 ± 2.045 | 1.223 ± 0.461 |
6.014 ± 0.891 | 6.116 ± 0.275 |
5.505 ± 0.497 | 7.034 ± 0.147 |
1.223 ± 0.08 | 3.364 ± 0.659 |
5.708 ± 0.843 | 7.441 ± 0.15 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.548 ± 0.577 | 2.956 ± 0.009 |
6.116 ± 0.266 | 3.466 ± 0.363 |
6.524 ± 0.384 | 9.48 ± 1.367 |
5.81 ± 0.043 | 6.116 ± 1.078 |
2.752 ± 0.564 | 3.16 ± 1.078 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
