
Lake Sarah-associated circular virus-30
Taxonomy: Viruses; unclassified viruses
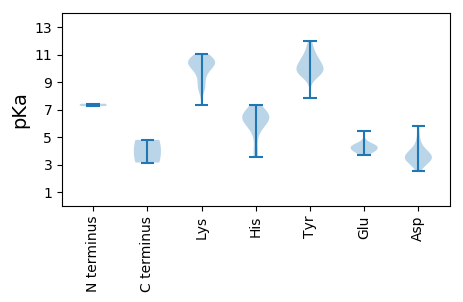
Average proteome isoelectric point is 7.66
Get precalculated fractions of proteins
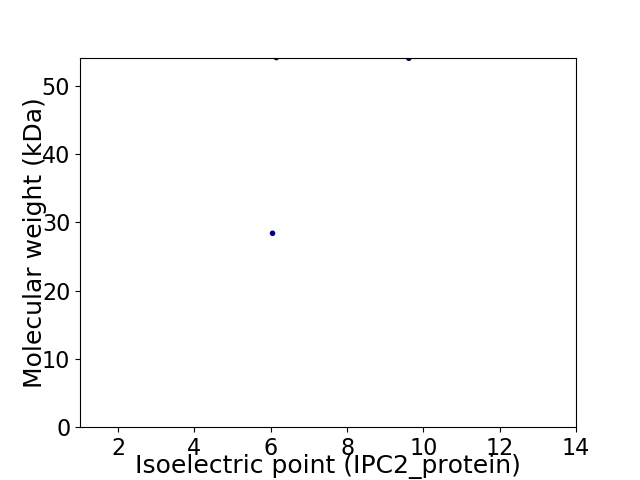
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQN3|A0A140AQN3_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-30 OX=1685758 PE=4 SV=1
MM1 pKa = 7.25SQGLYY6 pKa = 9.64WILTIPHH13 pKa = 7.33ADD15 pKa = 3.73FLPYY19 pKa = 10.37LPPGVQYY26 pKa = 10.81IRR28 pKa = 11.84GQLEE32 pKa = 3.71ASTLCRR38 pKa = 11.84PLADD42 pKa = 5.4DD43 pKa = 3.95YY44 pKa = 11.98GADD47 pKa = 3.29AGYY50 pKa = 10.03LHH52 pKa = 6.76WQLLVAFTRR61 pKa = 11.84KK62 pKa = 9.16CRR64 pKa = 11.84LAVVTRR70 pKa = 11.84TFGAGIHH77 pKa = 6.52AEE79 pKa = 4.25LSRR82 pKa = 11.84SSAANDD88 pKa = 3.69YY89 pKa = 9.65VWKK92 pKa = 10.24EE93 pKa = 3.96DD94 pKa = 3.4TRR96 pKa = 11.84VGGTQFEE103 pKa = 4.45LGKK106 pKa = 10.55LAFKK110 pKa = 10.74RR111 pKa = 11.84NSATDD116 pKa = 3.26WSSVRR121 pKa = 11.84DD122 pKa = 3.52SAKK125 pKa = 10.29RR126 pKa = 11.84GRR128 pKa = 11.84LDD130 pKa = 5.83DD131 pKa = 4.29IPSDD135 pKa = 3.24IYY137 pKa = 10.77VRR139 pKa = 11.84CYY141 pKa = 10.73NQLKK145 pKa = 10.45RR146 pKa = 11.84IACDD150 pKa = 3.22HH151 pKa = 5.79MQADD155 pKa = 3.67PMEE158 pKa = 5.46RR159 pKa = 11.84EE160 pKa = 4.56VVCYY164 pKa = 9.33WGRR167 pKa = 11.84SGSGKK172 pKa = 8.71SRR174 pKa = 11.84RR175 pKa = 11.84AWEE178 pKa = 3.89EE179 pKa = 4.03SGVNAYY185 pKa = 9.72PKK187 pKa = 10.39DD188 pKa = 3.75PRR190 pKa = 11.84SKK192 pKa = 10.25FWDD195 pKa = 4.41GYY197 pKa = 9.92RR198 pKa = 11.84GQEE201 pKa = 3.8HH202 pKa = 5.89VVLDD206 pKa = 4.06EE207 pKa = 4.0FRR209 pKa = 11.84GGIDD213 pKa = 2.92IAHH216 pKa = 6.28VLRR219 pKa = 11.84WFDD222 pKa = 3.68RR223 pKa = 11.84YY224 pKa = 9.64PVLEE228 pKa = 4.33GQGEE232 pKa = 4.79LGTLICKK239 pKa = 10.05EE240 pKa = 4.17DD241 pKa = 4.08LDD243 pKa = 4.94HH244 pKa = 6.58EE245 pKa = 4.47QHH247 pKa = 6.85SPP249 pKa = 3.14
MM1 pKa = 7.25SQGLYY6 pKa = 9.64WILTIPHH13 pKa = 7.33ADD15 pKa = 3.73FLPYY19 pKa = 10.37LPPGVQYY26 pKa = 10.81IRR28 pKa = 11.84GQLEE32 pKa = 3.71ASTLCRR38 pKa = 11.84PLADD42 pKa = 5.4DD43 pKa = 3.95YY44 pKa = 11.98GADD47 pKa = 3.29AGYY50 pKa = 10.03LHH52 pKa = 6.76WQLLVAFTRR61 pKa = 11.84KK62 pKa = 9.16CRR64 pKa = 11.84LAVVTRR70 pKa = 11.84TFGAGIHH77 pKa = 6.52AEE79 pKa = 4.25LSRR82 pKa = 11.84SSAANDD88 pKa = 3.69YY89 pKa = 9.65VWKK92 pKa = 10.24EE93 pKa = 3.96DD94 pKa = 3.4TRR96 pKa = 11.84VGGTQFEE103 pKa = 4.45LGKK106 pKa = 10.55LAFKK110 pKa = 10.74RR111 pKa = 11.84NSATDD116 pKa = 3.26WSSVRR121 pKa = 11.84DD122 pKa = 3.52SAKK125 pKa = 10.29RR126 pKa = 11.84GRR128 pKa = 11.84LDD130 pKa = 5.83DD131 pKa = 4.29IPSDD135 pKa = 3.24IYY137 pKa = 10.77VRR139 pKa = 11.84CYY141 pKa = 10.73NQLKK145 pKa = 10.45RR146 pKa = 11.84IACDD150 pKa = 3.22HH151 pKa = 5.79MQADD155 pKa = 3.67PMEE158 pKa = 5.46RR159 pKa = 11.84EE160 pKa = 4.56VVCYY164 pKa = 9.33WGRR167 pKa = 11.84SGSGKK172 pKa = 8.71SRR174 pKa = 11.84RR175 pKa = 11.84AWEE178 pKa = 3.89EE179 pKa = 4.03SGVNAYY185 pKa = 9.72PKK187 pKa = 10.39DD188 pKa = 3.75PRR190 pKa = 11.84SKK192 pKa = 10.25FWDD195 pKa = 4.41GYY197 pKa = 9.92RR198 pKa = 11.84GQEE201 pKa = 3.8HH202 pKa = 5.89VVLDD206 pKa = 4.06EE207 pKa = 4.0FRR209 pKa = 11.84GGIDD213 pKa = 2.92IAHH216 pKa = 6.28VLRR219 pKa = 11.84WFDD222 pKa = 3.68RR223 pKa = 11.84YY224 pKa = 9.64PVLEE228 pKa = 4.33GQGEE232 pKa = 4.79LGTLICKK239 pKa = 10.05EE240 pKa = 4.17DD241 pKa = 4.08LDD243 pKa = 4.94HH244 pKa = 6.58EE245 pKa = 4.47QHH247 pKa = 6.85SPP249 pKa = 3.14
Molecular weight: 28.4 kDa
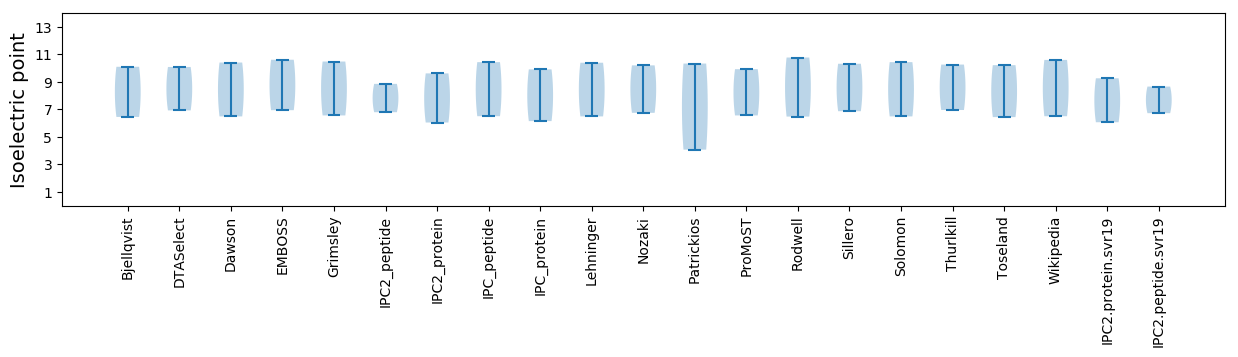
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQN3|A0A140AQN3_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-30 OX=1685758 PE=4 SV=1
MM1 pKa = 7.42PAVNGRR7 pKa = 11.84MTRR10 pKa = 11.84DD11 pKa = 2.56TWGDD15 pKa = 3.15HH16 pKa = 4.04FRR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 9.83YY21 pKa = 10.56GAASTMYY28 pKa = 9.63YY29 pKa = 9.93LHH31 pKa = 6.63NRR33 pKa = 11.84LPLSEE38 pKa = 4.06AQQRR42 pKa = 11.84KK43 pKa = 7.31AANWHH48 pKa = 6.09RR49 pKa = 11.84GKK51 pKa = 9.07KK52 pKa = 8.52TSSSQKK58 pKa = 10.27KK59 pKa = 8.33MSNGKK64 pKa = 9.62RR65 pKa = 11.84GRR67 pKa = 11.84SMSRR71 pKa = 11.84GRR73 pKa = 11.84SAKK76 pKa = 9.53RR77 pKa = 11.84RR78 pKa = 11.84PVTPRR83 pKa = 11.84RR84 pKa = 11.84TARR87 pKa = 11.84RR88 pKa = 11.84VSLPATPTARR98 pKa = 11.84SRR100 pKa = 11.84SVSRR104 pKa = 11.84GRR106 pKa = 11.84RR107 pKa = 11.84SARR110 pKa = 11.84SHH112 pKa = 5.13SVRR115 pKa = 11.84RR116 pKa = 11.84MPSLSRR122 pKa = 11.84SRR124 pKa = 11.84SAVSQPSASGSTGPSHH140 pKa = 6.6YY141 pKa = 10.45GSSARR146 pKa = 11.84GAGNLVVGTASSMLKK161 pKa = 10.12KK162 pKa = 8.63YY163 pKa = 10.82AKK165 pKa = 9.76YY166 pKa = 10.41GVVGEE171 pKa = 4.47DD172 pKa = 2.64QWTKK176 pKa = 10.66TIASTAAPIYY186 pKa = 10.21YY187 pKa = 9.98VVGHH191 pKa = 6.62CAPPNTPLLRR201 pKa = 11.84YY202 pKa = 9.15IAAATVKK209 pKa = 10.67AFFLQFGFGFDD220 pKa = 3.14NFTRR224 pKa = 11.84PLSDD228 pKa = 3.41YY229 pKa = 11.14GFTANTDD236 pKa = 3.5IIEE239 pKa = 4.37AQYY242 pKa = 9.43VTTPSGAATSFNIYY256 pKa = 9.63TYY258 pKa = 11.24VVGNTVGDD266 pKa = 3.77VVDD269 pKa = 3.95AFFVAFAGSSKK280 pKa = 10.38NLRR283 pKa = 11.84WDD285 pKa = 3.38FLKK288 pKa = 10.77FKK290 pKa = 10.8SVANPSSVHH299 pKa = 3.56VTMYY303 pKa = 10.4LQNAKK308 pKa = 9.38IHH310 pKa = 6.13LNSTSFLKK318 pKa = 10.67LQNRR322 pKa = 11.84TVEE325 pKa = 4.03SSADD329 pKa = 3.37IEE331 pKa = 4.32TDD333 pKa = 3.5DD334 pKa = 4.37VDD336 pKa = 3.82IMPLIGRR343 pKa = 11.84QFDD346 pKa = 3.55INGTILSLSNDD357 pKa = 3.97GYY359 pKa = 11.41QYY361 pKa = 11.75ACDD364 pKa = 3.42EE365 pKa = 4.39SFAKK369 pKa = 10.64GGLCTKK375 pKa = 10.66DD376 pKa = 2.87NGSAIKK382 pKa = 10.21AIPYY386 pKa = 7.87NAQVRR391 pKa = 11.84GKK393 pKa = 8.56STSKK397 pKa = 10.85NITVAPGEE405 pKa = 4.15ILTSKK410 pKa = 10.37IKK412 pKa = 10.41FEE414 pKa = 3.79KK415 pKa = 9.93TIYY418 pKa = 9.93FSKK421 pKa = 10.05LQSYY425 pKa = 10.1FDD427 pKa = 4.75NINGTGDD434 pKa = 2.86ISEE437 pKa = 4.4RR438 pKa = 11.84TWGKK442 pKa = 10.0SRR444 pKa = 11.84FFFFEE449 pKa = 4.07RR450 pKa = 11.84PIKK453 pKa = 10.78SLGDD457 pKa = 3.22GLAFNFKK464 pKa = 11.01AEE466 pKa = 4.25LDD468 pKa = 3.81VCHH471 pKa = 6.27EE472 pKa = 4.17VMVVGGNPPITTSTYY487 pKa = 10.05TIVAAA492 pKa = 4.79
MM1 pKa = 7.42PAVNGRR7 pKa = 11.84MTRR10 pKa = 11.84DD11 pKa = 2.56TWGDD15 pKa = 3.15HH16 pKa = 4.04FRR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 9.83YY21 pKa = 10.56GAASTMYY28 pKa = 9.63YY29 pKa = 9.93LHH31 pKa = 6.63NRR33 pKa = 11.84LPLSEE38 pKa = 4.06AQQRR42 pKa = 11.84KK43 pKa = 7.31AANWHH48 pKa = 6.09RR49 pKa = 11.84GKK51 pKa = 9.07KK52 pKa = 8.52TSSSQKK58 pKa = 10.27KK59 pKa = 8.33MSNGKK64 pKa = 9.62RR65 pKa = 11.84GRR67 pKa = 11.84SMSRR71 pKa = 11.84GRR73 pKa = 11.84SAKK76 pKa = 9.53RR77 pKa = 11.84RR78 pKa = 11.84PVTPRR83 pKa = 11.84RR84 pKa = 11.84TARR87 pKa = 11.84RR88 pKa = 11.84VSLPATPTARR98 pKa = 11.84SRR100 pKa = 11.84SVSRR104 pKa = 11.84GRR106 pKa = 11.84RR107 pKa = 11.84SARR110 pKa = 11.84SHH112 pKa = 5.13SVRR115 pKa = 11.84RR116 pKa = 11.84MPSLSRR122 pKa = 11.84SRR124 pKa = 11.84SAVSQPSASGSTGPSHH140 pKa = 6.6YY141 pKa = 10.45GSSARR146 pKa = 11.84GAGNLVVGTASSMLKK161 pKa = 10.12KK162 pKa = 8.63YY163 pKa = 10.82AKK165 pKa = 9.76YY166 pKa = 10.41GVVGEE171 pKa = 4.47DD172 pKa = 2.64QWTKK176 pKa = 10.66TIASTAAPIYY186 pKa = 10.21YY187 pKa = 9.98VVGHH191 pKa = 6.62CAPPNTPLLRR201 pKa = 11.84YY202 pKa = 9.15IAAATVKK209 pKa = 10.67AFFLQFGFGFDD220 pKa = 3.14NFTRR224 pKa = 11.84PLSDD228 pKa = 3.41YY229 pKa = 11.14GFTANTDD236 pKa = 3.5IIEE239 pKa = 4.37AQYY242 pKa = 9.43VTTPSGAATSFNIYY256 pKa = 9.63TYY258 pKa = 11.24VVGNTVGDD266 pKa = 3.77VVDD269 pKa = 3.95AFFVAFAGSSKK280 pKa = 10.38NLRR283 pKa = 11.84WDD285 pKa = 3.38FLKK288 pKa = 10.77FKK290 pKa = 10.8SVANPSSVHH299 pKa = 3.56VTMYY303 pKa = 10.4LQNAKK308 pKa = 9.38IHH310 pKa = 6.13LNSTSFLKK318 pKa = 10.67LQNRR322 pKa = 11.84TVEE325 pKa = 4.03SSADD329 pKa = 3.37IEE331 pKa = 4.32TDD333 pKa = 3.5DD334 pKa = 4.37VDD336 pKa = 3.82IMPLIGRR343 pKa = 11.84QFDD346 pKa = 3.55INGTILSLSNDD357 pKa = 3.97GYY359 pKa = 11.41QYY361 pKa = 11.75ACDD364 pKa = 3.42EE365 pKa = 4.39SFAKK369 pKa = 10.64GGLCTKK375 pKa = 10.66DD376 pKa = 2.87NGSAIKK382 pKa = 10.21AIPYY386 pKa = 7.87NAQVRR391 pKa = 11.84GKK393 pKa = 8.56STSKK397 pKa = 10.85NITVAPGEE405 pKa = 4.15ILTSKK410 pKa = 10.37IKK412 pKa = 10.41FEE414 pKa = 3.79KK415 pKa = 9.93TIYY418 pKa = 9.93FSKK421 pKa = 10.05LQSYY425 pKa = 10.1FDD427 pKa = 4.75NINGTGDD434 pKa = 2.86ISEE437 pKa = 4.4RR438 pKa = 11.84TWGKK442 pKa = 10.0SRR444 pKa = 11.84FFFFEE449 pKa = 4.07RR450 pKa = 11.84PIKK453 pKa = 10.78SLGDD457 pKa = 3.22GLAFNFKK464 pKa = 11.01AEE466 pKa = 4.25LDD468 pKa = 3.81VCHH471 pKa = 6.27EE472 pKa = 4.17VMVVGGNPPITTSTYY487 pKa = 10.05TIVAAA492 pKa = 4.79
Molecular weight: 54.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
741 |
249 |
492 |
370.5 |
41.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.637 ± 0.567 | 1.35 ± 0.598 |
5.533 ± 1.409 | 3.509 ± 1.191 |
4.588 ± 0.775 | 8.232 ± 0.34 |
2.294 ± 0.518 | 4.453 ± 0.247 |
5.263 ± 0.703 | 6.478 ± 1.329 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.754 ± 0.31 | 3.779 ± 1.224 |
4.453 ± 0.02 | 3.104 ± 0.514 |
7.962 ± 0.492 | 9.582 ± 1.779 |
6.478 ± 1.614 | 6.343 ± 0.18 |
1.754 ± 0.822 | 4.453 ± 0.206 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
