
Eubacterium hallii CAG:12
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriaceae; Eubacterium; environmental samples
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
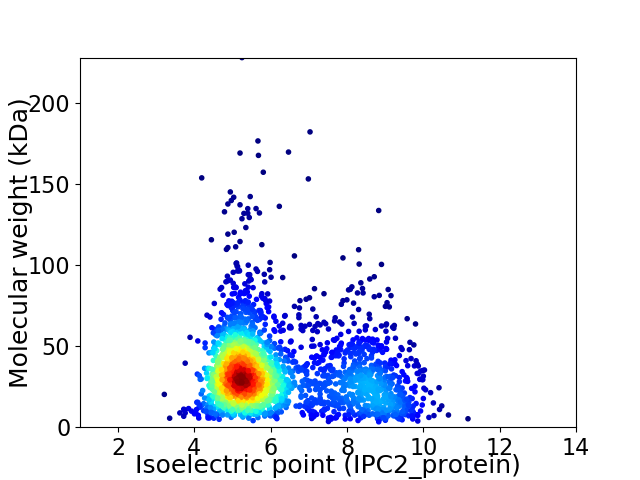
Virtual 2D-PAGE plot for 2309 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6GWN1|R6GWN1_9FIRM Carbohydrate kinase FGGY family protein OS=Eubacterium hallii CAG:12 OX=1263078 GN=BN476_01919 PE=4 SV=1
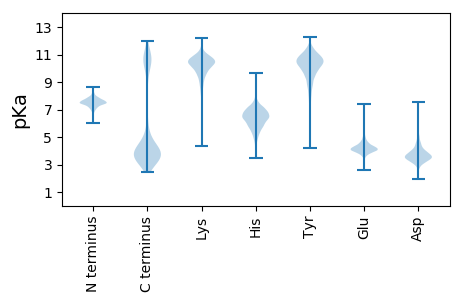
MM1 pKa = 7.56FKK3 pKa = 10.98NKK5 pKa = 9.74LKK7 pKa = 10.47RR8 pKa = 11.84LCAVTLAGMMVMSVSACSTSKK29 pKa = 10.56SAEE32 pKa = 3.99KK33 pKa = 10.78NSVSKK38 pKa = 11.23AEE40 pKa = 4.1QASEE44 pKa = 4.05ATTEE48 pKa = 3.93AAKK51 pKa = 10.8DD52 pKa = 3.64DD53 pKa = 3.81TTADD57 pKa = 3.72YY58 pKa = 10.81VASGVEE64 pKa = 4.01LPDD67 pKa = 3.34NFEE70 pKa = 4.0NMIYY74 pKa = 10.14PIEE77 pKa = 4.35ALMLQNYY84 pKa = 9.47SKK86 pKa = 10.97GYY88 pKa = 8.89PYY90 pKa = 10.21YY91 pKa = 11.17ANGASEE97 pKa = 4.5EE98 pKa = 4.32KK99 pKa = 10.41SDD101 pKa = 4.44SFWFSMAALTSLMEE115 pKa = 4.23NEE117 pKa = 4.07SAYY120 pKa = 10.81GDD122 pKa = 4.38GIEE125 pKa = 4.11MDD127 pKa = 3.16SYY129 pKa = 11.97YY130 pKa = 10.8YY131 pKa = 10.93LKK133 pKa = 10.89EE134 pKa = 4.03NTSNMYY140 pKa = 10.93ASALYY145 pKa = 9.87NAYY148 pKa = 10.44GMGNMEE154 pKa = 4.84FPEE157 pKa = 5.02IPDD160 pKa = 3.35GDD162 pKa = 3.74KK163 pKa = 11.08YY164 pKa = 10.96ATYY167 pKa = 10.99DD168 pKa = 4.61DD169 pKa = 4.19GKK171 pKa = 11.19QMYY174 pKa = 9.67GFLKK178 pKa = 10.84GEE180 pKa = 4.16VSSLVPYY187 pKa = 9.07ITNCVKK193 pKa = 10.81DD194 pKa = 3.87GSDD197 pKa = 4.16YY198 pKa = 10.88IITSQLRR205 pKa = 11.84DD206 pKa = 3.39KK207 pKa = 10.92DD208 pKa = 3.51TDD210 pKa = 3.83EE211 pKa = 4.41VKK213 pKa = 9.72GTYY216 pKa = 9.4EE217 pKa = 3.87VTITASSFDD226 pKa = 3.64GDD228 pKa = 4.52DD229 pKa = 3.16NCFAYY234 pKa = 10.46SATAIRR240 pKa = 11.84QIGEE244 pKa = 4.15TDD246 pKa = 3.61SEE248 pKa = 4.36NQSTSSEE255 pKa = 4.21SEE257 pKa = 3.96EE258 pKa = 4.2ASTDD262 pKa = 3.54STSEE266 pKa = 4.03NGDD269 pKa = 3.43STDD272 pKa = 4.1ANSSTDD278 pKa = 3.29STDD281 pKa = 3.22ATDD284 pKa = 3.78STDD287 pKa = 3.28STEE290 pKa = 6.19GISQDD295 pKa = 4.6DD296 pKa = 4.32ALSQAKK302 pKa = 9.96DD303 pKa = 3.72YY304 pKa = 11.04YY305 pKa = 11.44GEE307 pKa = 4.3DD308 pKa = 3.16AEE310 pKa = 4.38YY311 pKa = 10.51SYY313 pKa = 11.26KK314 pKa = 10.72KK315 pKa = 10.29QVTVGDD321 pKa = 3.51KK322 pKa = 10.09EE323 pKa = 4.44YY324 pKa = 11.32YY325 pKa = 10.11DD326 pKa = 4.55FSVSGDD332 pKa = 3.55NVSSTDD338 pKa = 3.25VLVSVDD344 pKa = 3.61GQDD347 pKa = 4.62VIGGTQNDD355 pKa = 3.84DD356 pKa = 3.78GSWSFDD362 pKa = 3.2QQ363 pKa = 5.08
MM1 pKa = 7.56FKK3 pKa = 10.98NKK5 pKa = 9.74LKK7 pKa = 10.47RR8 pKa = 11.84LCAVTLAGMMVMSVSACSTSKK29 pKa = 10.56SAEE32 pKa = 3.99KK33 pKa = 10.78NSVSKK38 pKa = 11.23AEE40 pKa = 4.1QASEE44 pKa = 4.05ATTEE48 pKa = 3.93AAKK51 pKa = 10.8DD52 pKa = 3.64DD53 pKa = 3.81TTADD57 pKa = 3.72YY58 pKa = 10.81VASGVEE64 pKa = 4.01LPDD67 pKa = 3.34NFEE70 pKa = 4.0NMIYY74 pKa = 10.14PIEE77 pKa = 4.35ALMLQNYY84 pKa = 9.47SKK86 pKa = 10.97GYY88 pKa = 8.89PYY90 pKa = 10.21YY91 pKa = 11.17ANGASEE97 pKa = 4.5EE98 pKa = 4.32KK99 pKa = 10.41SDD101 pKa = 4.44SFWFSMAALTSLMEE115 pKa = 4.23NEE117 pKa = 4.07SAYY120 pKa = 10.81GDD122 pKa = 4.38GIEE125 pKa = 4.11MDD127 pKa = 3.16SYY129 pKa = 11.97YY130 pKa = 10.8YY131 pKa = 10.93LKK133 pKa = 10.89EE134 pKa = 4.03NTSNMYY140 pKa = 10.93ASALYY145 pKa = 9.87NAYY148 pKa = 10.44GMGNMEE154 pKa = 4.84FPEE157 pKa = 5.02IPDD160 pKa = 3.35GDD162 pKa = 3.74KK163 pKa = 11.08YY164 pKa = 10.96ATYY167 pKa = 10.99DD168 pKa = 4.61DD169 pKa = 4.19GKK171 pKa = 11.19QMYY174 pKa = 9.67GFLKK178 pKa = 10.84GEE180 pKa = 4.16VSSLVPYY187 pKa = 9.07ITNCVKK193 pKa = 10.81DD194 pKa = 3.87GSDD197 pKa = 4.16YY198 pKa = 10.88IITSQLRR205 pKa = 11.84DD206 pKa = 3.39KK207 pKa = 10.92DD208 pKa = 3.51TDD210 pKa = 3.83EE211 pKa = 4.41VKK213 pKa = 9.72GTYY216 pKa = 9.4EE217 pKa = 3.87VTITASSFDD226 pKa = 3.64GDD228 pKa = 4.52DD229 pKa = 3.16NCFAYY234 pKa = 10.46SATAIRR240 pKa = 11.84QIGEE244 pKa = 4.15TDD246 pKa = 3.61SEE248 pKa = 4.36NQSTSSEE255 pKa = 4.21SEE257 pKa = 3.96EE258 pKa = 4.2ASTDD262 pKa = 3.54STSEE266 pKa = 4.03NGDD269 pKa = 3.43STDD272 pKa = 4.1ANSSTDD278 pKa = 3.29STDD281 pKa = 3.22ATDD284 pKa = 3.78STDD287 pKa = 3.28STEE290 pKa = 6.19GISQDD295 pKa = 4.6DD296 pKa = 4.32ALSQAKK302 pKa = 9.96DD303 pKa = 3.72YY304 pKa = 11.04YY305 pKa = 11.44GEE307 pKa = 4.3DD308 pKa = 3.16AEE310 pKa = 4.38YY311 pKa = 10.51SYY313 pKa = 11.26KK314 pKa = 10.72KK315 pKa = 10.29QVTVGDD321 pKa = 3.51KK322 pKa = 10.09EE323 pKa = 4.44YY324 pKa = 11.32YY325 pKa = 10.11DD326 pKa = 4.55FSVSGDD332 pKa = 3.55NVSSTDD338 pKa = 3.25VLVSVDD344 pKa = 3.61GQDD347 pKa = 4.62VIGGTQNDD355 pKa = 3.84DD356 pKa = 3.78GSWSFDD362 pKa = 3.2QQ363 pKa = 5.08
Molecular weight: 39.5 kDa
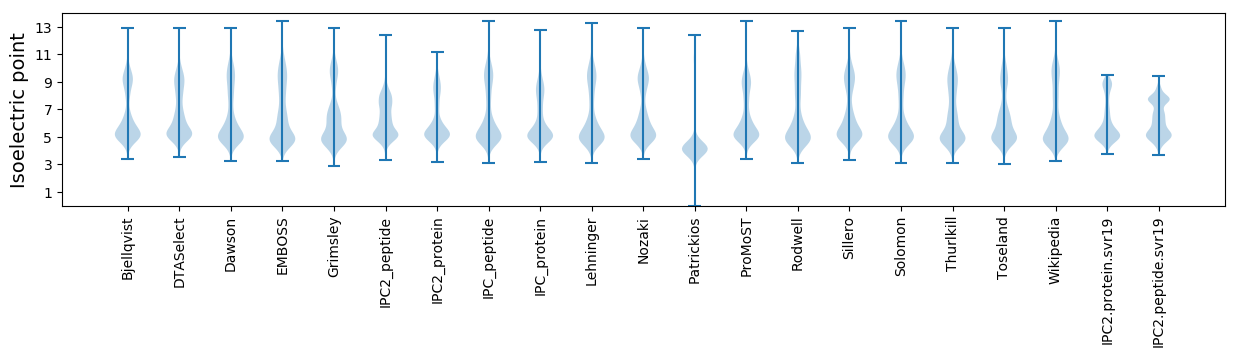
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6G9U3|R6G9U3_9FIRM Uncharacterized protein OS=Eubacterium hallii CAG:12 OX=1263078 GN=BN476_01577 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.58VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84NKK41 pKa = 10.44LSAA44 pKa = 3.84
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.95KK9 pKa = 7.58RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.07VHH16 pKa = 5.95GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.58VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84NKK41 pKa = 10.44LSAA44 pKa = 3.84
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
735250 |
29 |
2058 |
318.4 |
35.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.305 ± 0.056 | 1.514 ± 0.02 |
5.461 ± 0.041 | 7.857 ± 0.056 |
4.103 ± 0.034 | 7.004 ± 0.054 |
1.787 ± 0.024 | 7.728 ± 0.047 |
7.95 ± 0.051 | 8.64 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.084 ± 0.024 | 4.465 ± 0.035 |
3.155 ± 0.027 | 3.15 ± 0.033 |
4.002 ± 0.037 | 5.794 ± 0.042 |
5.363 ± 0.037 | 6.789 ± 0.044 |
0.781 ± 0.015 | 4.067 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
