
Kangiella geojedonensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Kangiellaceae; Kangiella
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
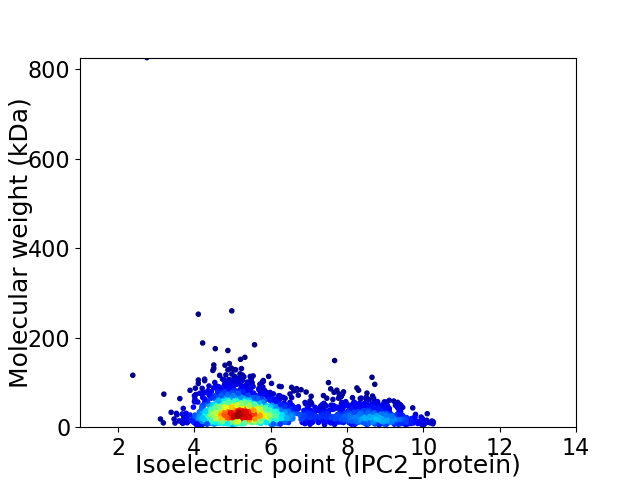
Virtual 2D-PAGE plot for 2207 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
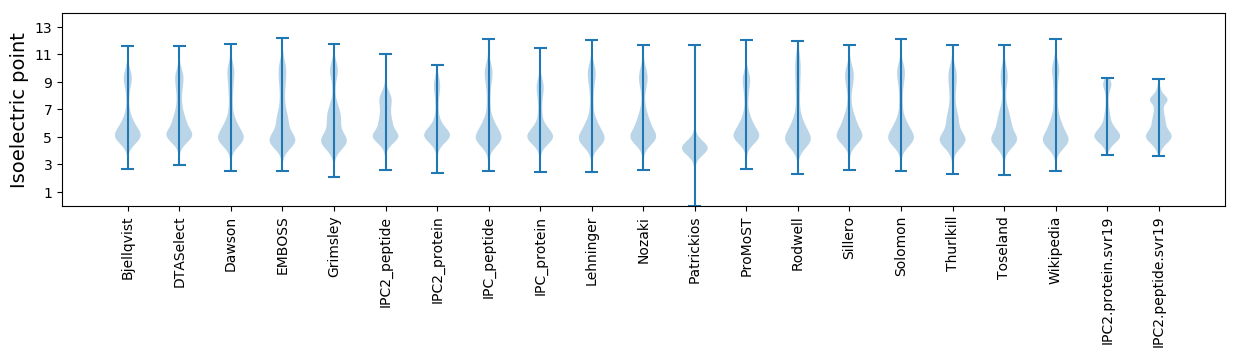
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F6RCU7|A0A0F6RCU7_9GAMM M6 family metalloprotease domain protein OS=Kangiella geojedonensis OX=914150 GN=TQ33_1379 PE=4 SV=1
MM1 pKa = 6.95VVSKK5 pKa = 10.74AVYY8 pKa = 9.84KK9 pKa = 10.8GFILLMLLLTCVRR22 pKa = 11.84VEE24 pKa = 4.03SATVEE29 pKa = 4.43FQVNWPEE36 pKa = 3.54WSQEE40 pKa = 3.74NRR42 pKa = 11.84IEE44 pKa = 4.38FYY46 pKa = 9.99NTAVDD51 pKa = 3.62VADD54 pKa = 4.82RR55 pKa = 11.84DD56 pKa = 3.81GDD58 pKa = 4.05GNTTEE63 pKa = 5.09IINSNLVTTRR73 pKa = 11.84YY74 pKa = 10.04GGEE77 pKa = 3.84NAICVPGFCTLPAVANTSFSQNYY100 pKa = 6.95TFNNVPSGNYY110 pKa = 9.48RR111 pKa = 11.84IVMYY115 pKa = 10.73DD116 pKa = 3.46DD117 pKa = 6.43FGDD120 pKa = 3.49GWNGGGTLTVLVNGTNIGTVTTTTSVTSTNVFTISDD156 pKa = 4.17TPTGTTAQTCSDD168 pKa = 4.08ANLTNAWSNLGASPQATAGAIPVSISTTDD197 pKa = 3.12TGPAGWVFSSDD208 pKa = 2.98TMNTINAWSSAAVQGRR224 pKa = 11.84TSLGVLFEE232 pKa = 4.38WDD234 pKa = 3.81PTPGANGFEE243 pKa = 4.27PVYY246 pKa = 10.52QATNSDD252 pKa = 3.56GVVNDD257 pKa = 4.01NDD259 pKa = 4.19PGGEE263 pKa = 3.97GTMDD267 pKa = 2.93ISFGGRR273 pKa = 11.84TVNNAIVHH281 pKa = 6.24IDD283 pKa = 3.26RR284 pKa = 11.84VGGIGAGVTNSSRR297 pKa = 11.84WGINTAGVTLGRR309 pKa = 11.84LAGVGHH315 pKa = 6.89FATYY319 pKa = 9.96TNNTFQRR326 pKa = 11.84DD327 pKa = 3.93MLVVSANTEE336 pKa = 4.06SSTNTATGTAAGSVVYY352 pKa = 10.29QGEE355 pKa = 4.42YY356 pKa = 9.17STLSLGLDD364 pKa = 4.24GIGLEE369 pKa = 4.48GAGGDD374 pKa = 3.67GMEE377 pKa = 4.58MVVCVPQADD386 pKa = 3.84LSLTKK391 pKa = 10.26TVDD394 pKa = 3.18NSNPNVGEE402 pKa = 4.0NVTYY406 pKa = 10.24TLTVTNNGPDD416 pKa = 3.48ATNDD420 pKa = 3.02IQVRR424 pKa = 11.84DD425 pKa = 4.18LLPAEE430 pKa = 4.7LDD432 pKa = 3.65FVSATEE438 pKa = 4.24SQGSFASGSGIWTVGTLANGASATLDD464 pKa = 3.02IVVSPNTAGVITNEE478 pKa = 3.95AEE480 pKa = 4.09IINADD485 pKa = 4.41RR486 pKa = 11.84IDD488 pKa = 4.16PDD490 pKa = 3.68SEE492 pKa = 4.12FRR494 pKa = 11.84SGLATDD500 pKa = 4.91DD501 pKa = 4.38LADD504 pKa = 5.51GIADD508 pKa = 4.05DD509 pKa = 5.78DD510 pKa = 4.3EE511 pKa = 6.79ASVDD515 pKa = 3.43INAGVVDD522 pKa = 3.9VSVTKK527 pKa = 10.7SSSNDD532 pKa = 3.73FIPGGTDD539 pKa = 2.74TYY541 pKa = 10.94TIIVSNAGPSDD552 pKa = 4.42AIGLLVSDD560 pKa = 4.86NLPSGVTLSGSWSCTPSAAGSSCNSASGGAAGGTSVSVTVNVAAGEE606 pKa = 4.21QVTITVPVEE615 pKa = 3.82YY616 pKa = 10.52SADD619 pKa = 3.59PTDD622 pKa = 3.67YY623 pKa = 11.62
MM1 pKa = 6.95VVSKK5 pKa = 10.74AVYY8 pKa = 9.84KK9 pKa = 10.8GFILLMLLLTCVRR22 pKa = 11.84VEE24 pKa = 4.03SATVEE29 pKa = 4.43FQVNWPEE36 pKa = 3.54WSQEE40 pKa = 3.74NRR42 pKa = 11.84IEE44 pKa = 4.38FYY46 pKa = 9.99NTAVDD51 pKa = 3.62VADD54 pKa = 4.82RR55 pKa = 11.84DD56 pKa = 3.81GDD58 pKa = 4.05GNTTEE63 pKa = 5.09IINSNLVTTRR73 pKa = 11.84YY74 pKa = 10.04GGEE77 pKa = 3.84NAICVPGFCTLPAVANTSFSQNYY100 pKa = 6.95TFNNVPSGNYY110 pKa = 9.48RR111 pKa = 11.84IVMYY115 pKa = 10.73DD116 pKa = 3.46DD117 pKa = 6.43FGDD120 pKa = 3.49GWNGGGTLTVLVNGTNIGTVTTTTSVTSTNVFTISDD156 pKa = 4.17TPTGTTAQTCSDD168 pKa = 4.08ANLTNAWSNLGASPQATAGAIPVSISTTDD197 pKa = 3.12TGPAGWVFSSDD208 pKa = 2.98TMNTINAWSSAAVQGRR224 pKa = 11.84TSLGVLFEE232 pKa = 4.38WDD234 pKa = 3.81PTPGANGFEE243 pKa = 4.27PVYY246 pKa = 10.52QATNSDD252 pKa = 3.56GVVNDD257 pKa = 4.01NDD259 pKa = 4.19PGGEE263 pKa = 3.97GTMDD267 pKa = 2.93ISFGGRR273 pKa = 11.84TVNNAIVHH281 pKa = 6.24IDD283 pKa = 3.26RR284 pKa = 11.84VGGIGAGVTNSSRR297 pKa = 11.84WGINTAGVTLGRR309 pKa = 11.84LAGVGHH315 pKa = 6.89FATYY319 pKa = 9.96TNNTFQRR326 pKa = 11.84DD327 pKa = 3.93MLVVSANTEE336 pKa = 4.06SSTNTATGTAAGSVVYY352 pKa = 10.29QGEE355 pKa = 4.42YY356 pKa = 9.17STLSLGLDD364 pKa = 4.24GIGLEE369 pKa = 4.48GAGGDD374 pKa = 3.67GMEE377 pKa = 4.58MVVCVPQADD386 pKa = 3.84LSLTKK391 pKa = 10.26TVDD394 pKa = 3.18NSNPNVGEE402 pKa = 4.0NVTYY406 pKa = 10.24TLTVTNNGPDD416 pKa = 3.48ATNDD420 pKa = 3.02IQVRR424 pKa = 11.84DD425 pKa = 4.18LLPAEE430 pKa = 4.7LDD432 pKa = 3.65FVSATEE438 pKa = 4.24SQGSFASGSGIWTVGTLANGASATLDD464 pKa = 3.02IVVSPNTAGVITNEE478 pKa = 3.95AEE480 pKa = 4.09IINADD485 pKa = 4.41RR486 pKa = 11.84IDD488 pKa = 4.16PDD490 pKa = 3.68SEE492 pKa = 4.12FRR494 pKa = 11.84SGLATDD500 pKa = 4.91DD501 pKa = 4.38LADD504 pKa = 5.51GIADD508 pKa = 4.05DD509 pKa = 5.78DD510 pKa = 4.3EE511 pKa = 6.79ASVDD515 pKa = 3.43INAGVVDD522 pKa = 3.9VSVTKK527 pKa = 10.7SSSNDD532 pKa = 3.73FIPGGTDD539 pKa = 2.74TYY541 pKa = 10.94TIIVSNAGPSDD552 pKa = 4.42AIGLLVSDD560 pKa = 4.86NLPSGVTLSGSWSCTPSAAGSSCNSASGGAAGGTSVSVTVNVAAGEE606 pKa = 4.21QVTITVPVEE615 pKa = 3.82YY616 pKa = 10.52SADD619 pKa = 3.59PTDD622 pKa = 3.67YY623 pKa = 11.62
Molecular weight: 63.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F6RAX0|A0A0F6RAX0_9GAMM 4-hydroxybenzoate octaprenyltransferase OS=Kangiella geojedonensis OX=914150 GN=ubiA PE=3 SV=1
MM1 pKa = 7.25ATINQLVRR9 pKa = 11.84KK10 pKa = 8.95PRR12 pKa = 11.84VKK14 pKa = 9.97QVKK17 pKa = 8.11KK18 pKa = 8.73TNVPALEE25 pKa = 4.4ACPQKK30 pKa = 10.67RR31 pKa = 11.84GVCTRR36 pKa = 11.84VYY38 pKa = 7.93TTTPKK43 pKa = 10.54KK44 pKa = 10.26PNSALRR50 pKa = 11.84KK51 pKa = 7.21VARR54 pKa = 11.84VRR56 pKa = 11.84LTNGYY61 pKa = 8.94EE62 pKa = 3.76VSSYY66 pKa = 10.86IGGEE70 pKa = 3.91GHH72 pKa = 6.6NLQEE76 pKa = 4.28HH77 pKa = 5.6SVVLIRR83 pKa = 11.84GGRR86 pKa = 11.84VKK88 pKa = 10.7DD89 pKa = 3.73LPGVRR94 pKa = 11.84YY95 pKa = 9.07HH96 pKa = 6.62CVRR99 pKa = 11.84GTLDD103 pKa = 3.37LAGVKK108 pKa = 9.49DD109 pKa = 4.14RR110 pKa = 11.84KK111 pKa = 9.86QGRR114 pKa = 11.84SKK116 pKa = 11.18YY117 pKa = 9.02GAKK120 pKa = 9.93RR121 pKa = 11.84PKK123 pKa = 10.33GG124 pKa = 3.5
MM1 pKa = 7.25ATINQLVRR9 pKa = 11.84KK10 pKa = 8.95PRR12 pKa = 11.84VKK14 pKa = 9.97QVKK17 pKa = 8.11KK18 pKa = 8.73TNVPALEE25 pKa = 4.4ACPQKK30 pKa = 10.67RR31 pKa = 11.84GVCTRR36 pKa = 11.84VYY38 pKa = 7.93TTTPKK43 pKa = 10.54KK44 pKa = 10.26PNSALRR50 pKa = 11.84KK51 pKa = 7.21VARR54 pKa = 11.84VRR56 pKa = 11.84LTNGYY61 pKa = 8.94EE62 pKa = 3.76VSSYY66 pKa = 10.86IGGEE70 pKa = 3.91GHH72 pKa = 6.6NLQEE76 pKa = 4.28HH77 pKa = 5.6SVVLIRR83 pKa = 11.84GGRR86 pKa = 11.84VKK88 pKa = 10.7DD89 pKa = 3.73LPGVRR94 pKa = 11.84YY95 pKa = 9.07HH96 pKa = 6.62CVRR99 pKa = 11.84GTLDD103 pKa = 3.37LAGVKK108 pKa = 9.49DD109 pKa = 4.14RR110 pKa = 11.84KK111 pKa = 9.86QGRR114 pKa = 11.84SKK116 pKa = 11.18YY117 pKa = 9.02GAKK120 pKa = 9.93RR121 pKa = 11.84PKK123 pKa = 10.33GG124 pKa = 3.5
Molecular weight: 13.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
737553 |
50 |
8155 |
334.2 |
37.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.116 ± 0.065 | 0.869 ± 0.021 |
6.073 ± 0.087 | 6.873 ± 0.061 |
4.066 ± 0.035 | 6.979 ± 0.057 |
2.198 ± 0.031 | 6.317 ± 0.041 |
5.893 ± 0.082 | 9.917 ± 0.083 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.448 ± 0.035 | 4.283 ± 0.042 |
3.867 ± 0.026 | 4.361 ± 0.049 |
4.353 ± 0.057 | 6.698 ± 0.054 |
5.341 ± 0.171 | 6.927 ± 0.047 |
1.211 ± 0.023 | 3.212 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
