
Influenza A virus (A/mallard/MN/51/1998(H2N3))
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Insthoviricetes; Articulavirales; Orthomyxoviridae; Alphainfluenzavirus; Influenza A virus; H2N3 subtype
Average proteome isoelectric point is 7.39
Get precalculated fractions of proteins
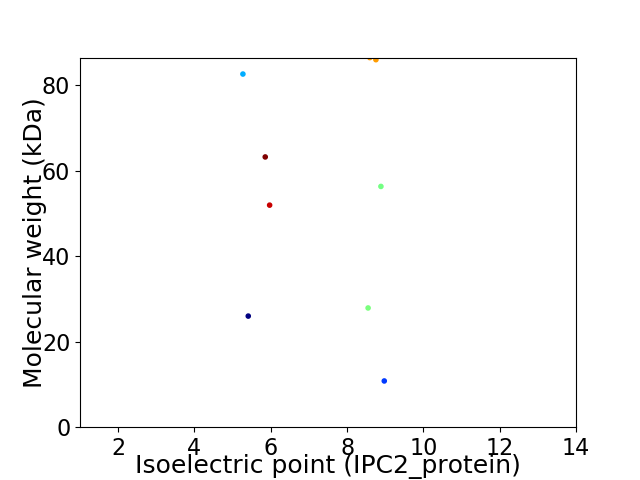
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B6EES3|B6EES3_9INFA RNA-directed RNA polymerase catalytic subunit OS=Influenza A virus (A/mallard/MN/51/1998(H2N3)) OX=566259 GN=PB1 PE=3 SV=1
MM1 pKa = 7.82EE2 pKa = 5.88DD3 pKa = 4.23FVRR6 pKa = 11.84QCFNPMIVEE15 pKa = 4.33LAEE18 pKa = 4.23KK19 pKa = 10.56AMKK22 pKa = 10.25EE23 pKa = 3.99YY24 pKa = 11.35GEE26 pKa = 4.4DD27 pKa = 3.65PKK29 pKa = 10.9IEE31 pKa = 4.27TNKK34 pKa = 9.66FAAICTHH41 pKa = 6.8LEE43 pKa = 3.9VCFMYY48 pKa = 10.77SDD50 pKa = 3.5FHH52 pKa = 8.77FIDD55 pKa = 3.6EE56 pKa = 4.51RR57 pKa = 11.84GEE59 pKa = 4.25SIIVEE64 pKa = 4.23SGDD67 pKa = 3.45PNALLKK73 pKa = 10.77HH74 pKa = 5.93RR75 pKa = 11.84FEE77 pKa = 6.44IIEE80 pKa = 4.15GRR82 pKa = 11.84DD83 pKa = 2.82RR84 pKa = 11.84TMAWTVVNSICNTTGVEE101 pKa = 4.2KK102 pKa = 10.67PKK104 pKa = 10.45FLPDD108 pKa = 3.39LYY110 pKa = 10.76DD111 pKa = 3.44YY112 pKa = 11.29KK113 pKa = 11.02EE114 pKa = 3.78NRR116 pKa = 11.84FIEE119 pKa = 4.17IGVTRR124 pKa = 11.84RR125 pKa = 11.84EE126 pKa = 3.6VHH128 pKa = 6.1IYY130 pKa = 10.32YY131 pKa = 10.35LEE133 pKa = 4.16KK134 pKa = 10.68ANKK137 pKa = 8.54IKK139 pKa = 10.7SEE141 pKa = 3.97KK142 pKa = 7.72THH144 pKa = 6.1IHH146 pKa = 5.76IFSFTGEE153 pKa = 3.88EE154 pKa = 4.15MATKK158 pKa = 10.42ADD160 pKa = 3.63YY161 pKa = 10.43TLDD164 pKa = 3.45EE165 pKa = 4.41EE166 pKa = 4.38SRR168 pKa = 11.84ARR170 pKa = 11.84IKK172 pKa = 10.03TRR174 pKa = 11.84LFTIRR179 pKa = 11.84QEE181 pKa = 4.07MASRR185 pKa = 11.84GLWDD189 pKa = 4.1PFRR192 pKa = 11.84QSEE195 pKa = 4.24RR196 pKa = 11.84GEE198 pKa = 3.89EE199 pKa = 4.16TIEE202 pKa = 3.68EE203 pKa = 4.17RR204 pKa = 11.84FEE206 pKa = 3.72ITGTMRR212 pKa = 11.84RR213 pKa = 11.84LADD216 pKa = 3.3QSLPPNFSSLEE227 pKa = 3.83NFRR230 pKa = 11.84AYY232 pKa = 11.05VDD234 pKa = 3.5GFEE237 pKa = 4.68PNGCIEE243 pKa = 4.47GKK245 pKa = 9.96LSQMSKK251 pKa = 9.86EE252 pKa = 3.84VNARR256 pKa = 11.84IEE258 pKa = 4.38PFLKK262 pKa = 8.15TTPRR266 pKa = 11.84PLRR269 pKa = 11.84LPEE272 pKa = 4.79GPPCSQRR279 pKa = 11.84SKK281 pKa = 10.91FLLMDD286 pKa = 4.4ALKK289 pKa = 10.88LSIEE293 pKa = 4.56DD294 pKa = 4.07PSHH297 pKa = 6.12EE298 pKa = 4.34GEE300 pKa = 5.39GIPLYY305 pKa = 10.72DD306 pKa = 5.54AIKK309 pKa = 10.45CMKK312 pKa = 9.82TFFGWKK318 pKa = 8.64EE319 pKa = 3.72PNIVKK324 pKa = 9.51PHH326 pKa = 6.17EE327 pKa = 4.45KK328 pKa = 10.38GINPNYY334 pKa = 9.91LLAWKK339 pKa = 9.87QVLAEE344 pKa = 4.25LQDD347 pKa = 3.52IEE349 pKa = 4.52NEE351 pKa = 4.07EE352 pKa = 4.77KK353 pKa = 10.44IPKK356 pKa = 7.57TKK358 pKa = 10.66NMKK361 pKa = 8.74KK362 pKa = 8.8TSQLKK367 pKa = 8.56WALGEE372 pKa = 3.97NMAPEE377 pKa = 4.21KK378 pKa = 10.99VDD380 pKa = 5.68FEE382 pKa = 4.64DD383 pKa = 4.93CKK385 pKa = 11.16DD386 pKa = 3.67VSDD389 pKa = 4.28LRR391 pKa = 11.84QYY393 pKa = 11.66DD394 pKa = 3.41SDD396 pKa = 3.88EE397 pKa = 4.32PEE399 pKa = 4.05QRR401 pKa = 11.84SLASWIQSEE410 pKa = 4.32FNKK413 pKa = 10.5ACEE416 pKa = 4.41LTDD419 pKa = 3.77SSWIEE424 pKa = 3.62LDD426 pKa = 4.1EE427 pKa = 5.66IGEE430 pKa = 4.15DD431 pKa = 3.56VAPIEE436 pKa = 5.34HH437 pKa = 6.74IASMRR442 pKa = 11.84RR443 pKa = 11.84NYY445 pKa = 8.17FTAEE449 pKa = 3.92VSHH452 pKa = 6.6CRR454 pKa = 11.84ATEE457 pKa = 3.87YY458 pKa = 10.63IMKK461 pKa = 10.18GVYY464 pKa = 10.04INTALLNASCAAMDD478 pKa = 4.67DD479 pKa = 4.09FQLIPMISKK488 pKa = 10.11CRR490 pKa = 11.84TKK492 pKa = 10.42EE493 pKa = 3.77GRR495 pKa = 11.84RR496 pKa = 11.84KK497 pKa = 7.6TNLYY501 pKa = 10.52GFIIKK506 pKa = 9.94GRR508 pKa = 11.84SHH510 pKa = 7.02LRR512 pKa = 11.84NDD514 pKa = 3.25TDD516 pKa = 3.55VVNFVSMEE524 pKa = 4.12FSLTDD529 pKa = 3.59PRR531 pKa = 11.84LEE533 pKa = 3.7PHH535 pKa = 6.34KK536 pKa = 10.03WEE538 pKa = 4.59KK539 pKa = 11.01YY540 pKa = 8.74CVLEE544 pKa = 5.19IGDD547 pKa = 4.0MLLRR551 pKa = 11.84TAIGQVSRR559 pKa = 11.84PMFLYY564 pKa = 10.52VRR566 pKa = 11.84TNGTSKK572 pKa = 10.61IKK574 pKa = 9.96MKK576 pKa = 9.82WGMEE580 pKa = 3.71MRR582 pKa = 11.84RR583 pKa = 11.84CLLQSLQQIEE593 pKa = 4.49SMIEE597 pKa = 3.69AEE599 pKa = 4.34SSVKK603 pKa = 10.34EE604 pKa = 3.72KK605 pKa = 11.43DD606 pKa = 3.06MTKK609 pKa = 10.59EE610 pKa = 3.6FFEE613 pKa = 4.76NKK615 pKa = 9.96SEE617 pKa = 3.88TWPIGEE623 pKa = 4.22SPKK626 pKa = 10.75GVEE629 pKa = 4.03EE630 pKa = 4.32GSIGKK635 pKa = 8.56VCRR638 pKa = 11.84TLLAKK643 pKa = 10.3SVFNSLYY650 pKa = 10.64ASPQLEE656 pKa = 4.23GFSAEE661 pKa = 4.07SRR663 pKa = 11.84KK664 pKa = 10.13LLLIVQALMDD674 pKa = 4.2NLEE677 pKa = 4.82PGTFDD682 pKa = 5.09LGGLYY687 pKa = 9.85EE688 pKa = 5.25AIEE691 pKa = 4.38EE692 pKa = 4.37CLINDD697 pKa = 3.63PWVLLNASWFNSFLTHH713 pKa = 6.92ALKK716 pKa = 10.78
MM1 pKa = 7.82EE2 pKa = 5.88DD3 pKa = 4.23FVRR6 pKa = 11.84QCFNPMIVEE15 pKa = 4.33LAEE18 pKa = 4.23KK19 pKa = 10.56AMKK22 pKa = 10.25EE23 pKa = 3.99YY24 pKa = 11.35GEE26 pKa = 4.4DD27 pKa = 3.65PKK29 pKa = 10.9IEE31 pKa = 4.27TNKK34 pKa = 9.66FAAICTHH41 pKa = 6.8LEE43 pKa = 3.9VCFMYY48 pKa = 10.77SDD50 pKa = 3.5FHH52 pKa = 8.77FIDD55 pKa = 3.6EE56 pKa = 4.51RR57 pKa = 11.84GEE59 pKa = 4.25SIIVEE64 pKa = 4.23SGDD67 pKa = 3.45PNALLKK73 pKa = 10.77HH74 pKa = 5.93RR75 pKa = 11.84FEE77 pKa = 6.44IIEE80 pKa = 4.15GRR82 pKa = 11.84DD83 pKa = 2.82RR84 pKa = 11.84TMAWTVVNSICNTTGVEE101 pKa = 4.2KK102 pKa = 10.67PKK104 pKa = 10.45FLPDD108 pKa = 3.39LYY110 pKa = 10.76DD111 pKa = 3.44YY112 pKa = 11.29KK113 pKa = 11.02EE114 pKa = 3.78NRR116 pKa = 11.84FIEE119 pKa = 4.17IGVTRR124 pKa = 11.84RR125 pKa = 11.84EE126 pKa = 3.6VHH128 pKa = 6.1IYY130 pKa = 10.32YY131 pKa = 10.35LEE133 pKa = 4.16KK134 pKa = 10.68ANKK137 pKa = 8.54IKK139 pKa = 10.7SEE141 pKa = 3.97KK142 pKa = 7.72THH144 pKa = 6.1IHH146 pKa = 5.76IFSFTGEE153 pKa = 3.88EE154 pKa = 4.15MATKK158 pKa = 10.42ADD160 pKa = 3.63YY161 pKa = 10.43TLDD164 pKa = 3.45EE165 pKa = 4.41EE166 pKa = 4.38SRR168 pKa = 11.84ARR170 pKa = 11.84IKK172 pKa = 10.03TRR174 pKa = 11.84LFTIRR179 pKa = 11.84QEE181 pKa = 4.07MASRR185 pKa = 11.84GLWDD189 pKa = 4.1PFRR192 pKa = 11.84QSEE195 pKa = 4.24RR196 pKa = 11.84GEE198 pKa = 3.89EE199 pKa = 4.16TIEE202 pKa = 3.68EE203 pKa = 4.17RR204 pKa = 11.84FEE206 pKa = 3.72ITGTMRR212 pKa = 11.84RR213 pKa = 11.84LADD216 pKa = 3.3QSLPPNFSSLEE227 pKa = 3.83NFRR230 pKa = 11.84AYY232 pKa = 11.05VDD234 pKa = 3.5GFEE237 pKa = 4.68PNGCIEE243 pKa = 4.47GKK245 pKa = 9.96LSQMSKK251 pKa = 9.86EE252 pKa = 3.84VNARR256 pKa = 11.84IEE258 pKa = 4.38PFLKK262 pKa = 8.15TTPRR266 pKa = 11.84PLRR269 pKa = 11.84LPEE272 pKa = 4.79GPPCSQRR279 pKa = 11.84SKK281 pKa = 10.91FLLMDD286 pKa = 4.4ALKK289 pKa = 10.88LSIEE293 pKa = 4.56DD294 pKa = 4.07PSHH297 pKa = 6.12EE298 pKa = 4.34GEE300 pKa = 5.39GIPLYY305 pKa = 10.72DD306 pKa = 5.54AIKK309 pKa = 10.45CMKK312 pKa = 9.82TFFGWKK318 pKa = 8.64EE319 pKa = 3.72PNIVKK324 pKa = 9.51PHH326 pKa = 6.17EE327 pKa = 4.45KK328 pKa = 10.38GINPNYY334 pKa = 9.91LLAWKK339 pKa = 9.87QVLAEE344 pKa = 4.25LQDD347 pKa = 3.52IEE349 pKa = 4.52NEE351 pKa = 4.07EE352 pKa = 4.77KK353 pKa = 10.44IPKK356 pKa = 7.57TKK358 pKa = 10.66NMKK361 pKa = 8.74KK362 pKa = 8.8TSQLKK367 pKa = 8.56WALGEE372 pKa = 3.97NMAPEE377 pKa = 4.21KK378 pKa = 10.99VDD380 pKa = 5.68FEE382 pKa = 4.64DD383 pKa = 4.93CKK385 pKa = 11.16DD386 pKa = 3.67VSDD389 pKa = 4.28LRR391 pKa = 11.84QYY393 pKa = 11.66DD394 pKa = 3.41SDD396 pKa = 3.88EE397 pKa = 4.32PEE399 pKa = 4.05QRR401 pKa = 11.84SLASWIQSEE410 pKa = 4.32FNKK413 pKa = 10.5ACEE416 pKa = 4.41LTDD419 pKa = 3.77SSWIEE424 pKa = 3.62LDD426 pKa = 4.1EE427 pKa = 5.66IGEE430 pKa = 4.15DD431 pKa = 3.56VAPIEE436 pKa = 5.34HH437 pKa = 6.74IASMRR442 pKa = 11.84RR443 pKa = 11.84NYY445 pKa = 8.17FTAEE449 pKa = 3.92VSHH452 pKa = 6.6CRR454 pKa = 11.84ATEE457 pKa = 3.87YY458 pKa = 10.63IMKK461 pKa = 10.18GVYY464 pKa = 10.04INTALLNASCAAMDD478 pKa = 4.67DD479 pKa = 4.09FQLIPMISKK488 pKa = 10.11CRR490 pKa = 11.84TKK492 pKa = 10.42EE493 pKa = 3.77GRR495 pKa = 11.84RR496 pKa = 11.84KK497 pKa = 7.6TNLYY501 pKa = 10.52GFIIKK506 pKa = 9.94GRR508 pKa = 11.84SHH510 pKa = 7.02LRR512 pKa = 11.84NDD514 pKa = 3.25TDD516 pKa = 3.55VVNFVSMEE524 pKa = 4.12FSLTDD529 pKa = 3.59PRR531 pKa = 11.84LEE533 pKa = 3.7PHH535 pKa = 6.34KK536 pKa = 10.03WEE538 pKa = 4.59KK539 pKa = 11.01YY540 pKa = 8.74CVLEE544 pKa = 5.19IGDD547 pKa = 4.0MLLRR551 pKa = 11.84TAIGQVSRR559 pKa = 11.84PMFLYY564 pKa = 10.52VRR566 pKa = 11.84TNGTSKK572 pKa = 10.61IKK574 pKa = 9.96MKK576 pKa = 9.82WGMEE580 pKa = 3.71MRR582 pKa = 11.84RR583 pKa = 11.84CLLQSLQQIEE593 pKa = 4.49SMIEE597 pKa = 3.69AEE599 pKa = 4.34SSVKK603 pKa = 10.34EE604 pKa = 3.72KK605 pKa = 11.43DD606 pKa = 3.06MTKK609 pKa = 10.59EE610 pKa = 3.6FFEE613 pKa = 4.76NKK615 pKa = 9.96SEE617 pKa = 3.88TWPIGEE623 pKa = 4.22SPKK626 pKa = 10.75GVEE629 pKa = 4.03EE630 pKa = 4.32GSIGKK635 pKa = 8.56VCRR638 pKa = 11.84TLLAKK643 pKa = 10.3SVFNSLYY650 pKa = 10.64ASPQLEE656 pKa = 4.23GFSAEE661 pKa = 4.07SRR663 pKa = 11.84KK664 pKa = 10.13LLLIVQALMDD674 pKa = 4.2NLEE677 pKa = 4.82PGTFDD682 pKa = 5.09LGGLYY687 pKa = 9.85EE688 pKa = 5.25AIEE691 pKa = 4.38EE692 pKa = 4.37CLINDD697 pKa = 3.63PWVLLNASWFNSFLTHH713 pKa = 6.92ALKK716 pKa = 10.78
Molecular weight: 82.58 kDa
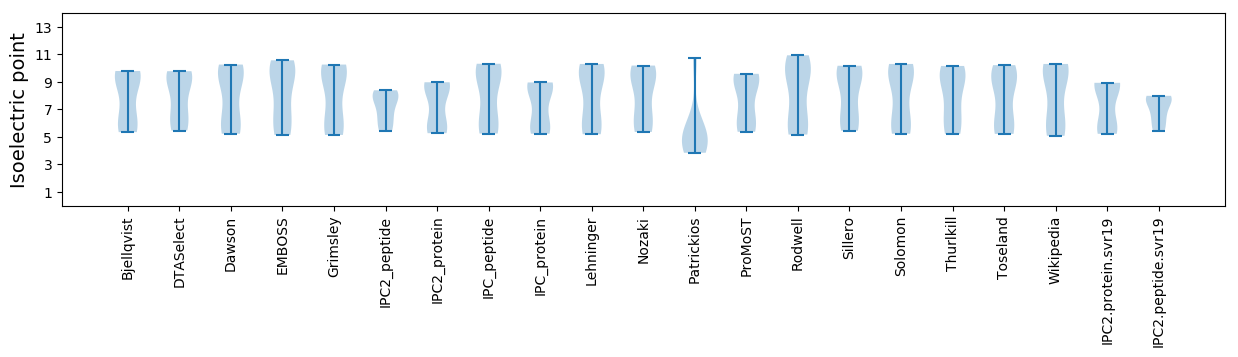
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B6EES5|B6EES5_9INFA Polymerase basic protein 2 OS=Influenza A virus (A/mallard/MN/51/1998(H2N3)) OX=566259 GN=PB2 PE=3 SV=1
MM1 pKa = 7.42EE2 pKa = 5.8RR3 pKa = 11.84EE4 pKa = 3.77QDD6 pKa = 4.01TPWTQSTEE14 pKa = 4.0HH15 pKa = 7.17INIQKK20 pKa = 10.18RR21 pKa = 11.84GNGQPTQRR29 pKa = 11.84LEE31 pKa = 4.29HH32 pKa = 6.81PNLTQLTDD40 pKa = 2.94HH41 pKa = 7.36CLRR44 pKa = 11.84TMSQVDD50 pKa = 3.29MHH52 pKa = 6.48KK53 pKa = 9.27QTVSLKK59 pKa = 9.82QWLSSKK65 pKa = 10.89NPTQEE70 pKa = 3.97SLKK73 pKa = 9.98TRR75 pKa = 11.84VLKK78 pKa = 10.36RR79 pKa = 11.84WKK81 pKa = 10.33LFNKK85 pKa = 9.41QEE87 pKa = 4.01WTNN90 pKa = 3.4
MM1 pKa = 7.42EE2 pKa = 5.8RR3 pKa = 11.84EE4 pKa = 3.77QDD6 pKa = 4.01TPWTQSTEE14 pKa = 4.0HH15 pKa = 7.17INIQKK20 pKa = 10.18RR21 pKa = 11.84GNGQPTQRR29 pKa = 11.84LEE31 pKa = 4.29HH32 pKa = 6.81PNLTQLTDD40 pKa = 2.94HH41 pKa = 7.36CLRR44 pKa = 11.84TMSQVDD50 pKa = 3.29MHH52 pKa = 6.48KK53 pKa = 9.27QTVSLKK59 pKa = 9.82QWLSSKK65 pKa = 10.89NPTQEE70 pKa = 3.97SLKK73 pKa = 9.98TRR75 pKa = 11.84VLKK78 pKa = 10.36RR79 pKa = 11.84WKK81 pKa = 10.33LFNKK85 pKa = 9.41QEE87 pKa = 4.01WTNN90 pKa = 3.4
Molecular weight: 10.81 kDa
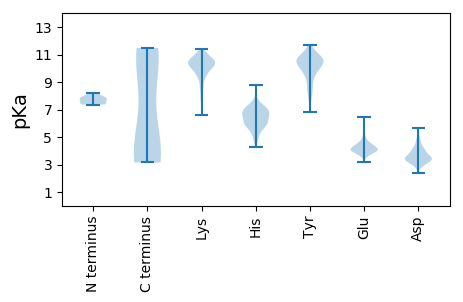
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4333 |
90 |
759 |
481.4 |
54.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.724 ± 0.526 | 1.777 ± 0.4 |
4.662 ± 0.289 | 7.385 ± 0.744 |
3.785 ± 0.288 | 6.67 ± 0.464 |
1.708 ± 0.184 | 6.531 ± 0.44 |
5.977 ± 0.464 | 7.824 ± 0.499 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.9 ± 0.48 | 5.562 ± 0.484 |
4.108 ± 0.296 | 3.877 ± 0.466 |
6.739 ± 0.794 | 6.993 ± 0.192 |
6.97 ± 0.442 | 5.608 ± 0.513 |
1.592 ± 0.19 | 2.608 ± 0.228 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
