
Blueberry shock virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Ilarvirus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
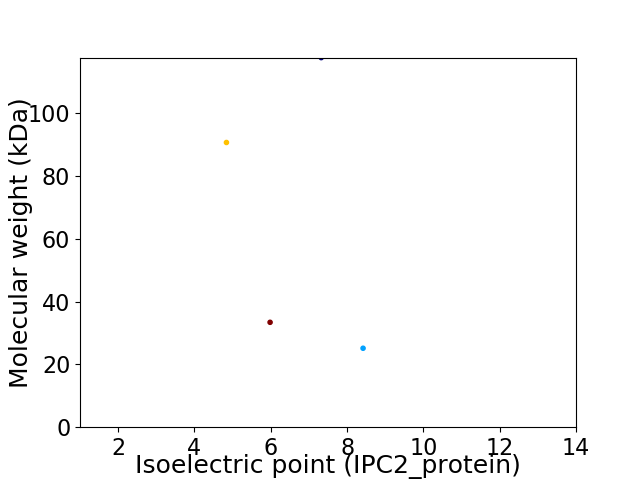
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T1WFS1|T1WFS1_9BROM RNA-directed RNA polymerase 2a OS=Blueberry shock virus OX=747056 PE=4 SV=1
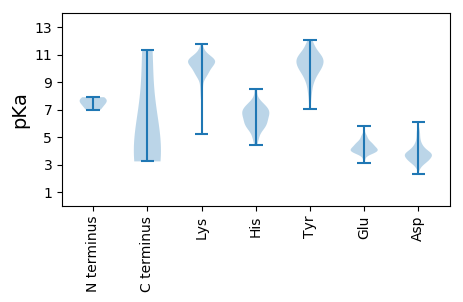
MM1 pKa = 7.68AFLSSMFPSDD11 pKa = 4.17SFLDD15 pKa = 4.0RR16 pKa = 11.84DD17 pKa = 3.75WEE19 pKa = 4.53VPLLLDD25 pKa = 3.5EE26 pKa = 4.74RR27 pKa = 11.84PRR29 pKa = 11.84TEE31 pKa = 3.88VEE33 pKa = 3.92QVSNEE38 pKa = 4.06EE39 pKa = 3.79VDD41 pKa = 3.78TNALFLKK48 pKa = 10.43SYY50 pKa = 10.87LLTLFLSPKK59 pKa = 9.74FKK61 pKa = 11.17YY62 pKa = 10.14IFEE65 pKa = 5.1DD66 pKa = 4.0FLGTEE71 pKa = 4.58FACYY75 pKa = 9.86VKK77 pKa = 10.34PRR79 pKa = 11.84WAIVFGCEE87 pKa = 3.14VDD89 pKa = 4.05YY90 pKa = 10.99EE91 pKa = 4.35VHH93 pKa = 6.4EE94 pKa = 4.46LVNDD98 pKa = 3.92EE99 pKa = 5.22LPLHH103 pKa = 6.88LDD105 pKa = 3.31TYY107 pKa = 9.87EE108 pKa = 3.94PWEE111 pKa = 4.34GVDD114 pKa = 6.1DD115 pKa = 4.68EE116 pKa = 4.97PCLGIDD122 pKa = 4.99DD123 pKa = 5.48LLDD126 pKa = 4.28RR127 pKa = 11.84DD128 pKa = 3.91ASPVEE133 pKa = 3.89RR134 pKa = 11.84DD135 pKa = 3.5DD136 pKa = 5.57VLFCLEE142 pKa = 4.92DD143 pKa = 3.81EE144 pKa = 4.63SDD146 pKa = 3.71GTTSLVLPTQTGIDD160 pKa = 4.17DD161 pKa = 5.36DD162 pKa = 4.66VDD164 pKa = 4.25SDD166 pKa = 3.77TSLKK170 pKa = 10.59VFEE173 pKa = 4.38STEE176 pKa = 3.92DD177 pKa = 3.78LEE179 pKa = 5.23DD180 pKa = 3.48VKK182 pKa = 10.53PTLKK186 pKa = 10.89SSDD189 pKa = 3.38LVLRR193 pKa = 11.84TINDD197 pKa = 3.25VDD199 pKa = 3.81TVIPVVEE206 pKa = 4.25KK207 pKa = 10.57FDD209 pKa = 3.91TNRR212 pKa = 11.84LHH214 pKa = 6.88LVKK217 pKa = 10.14EE218 pKa = 4.04VMMDD222 pKa = 3.5GFEE225 pKa = 4.43SLLIAHH231 pKa = 6.33EE232 pKa = 5.2RR233 pKa = 11.84ILSRR237 pKa = 11.84NDD239 pKa = 2.97WASLAMYY246 pKa = 9.12CPDD249 pKa = 4.33MEE251 pKa = 5.74IPTCGDD257 pKa = 3.35LLEE260 pKa = 5.48IPVKK264 pKa = 9.95HH265 pKa = 6.24TDD267 pKa = 2.83IEE269 pKa = 4.72FIQMAVDD276 pKa = 3.45EE277 pKa = 5.08LLPGVCDD284 pKa = 4.24LNDD287 pKa = 3.6SFFQEE292 pKa = 4.49LVEE295 pKa = 4.45TGDD298 pKa = 3.85IALEE302 pKa = 3.9LDD304 pKa = 3.15KK305 pKa = 11.57AKK307 pKa = 10.18IDD309 pKa = 3.2RR310 pKa = 11.84SVFNDD315 pKa = 2.81WSKK318 pKa = 11.44SKK320 pKa = 10.63KK321 pKa = 10.12CISSRR326 pKa = 11.84LNTGNISKK334 pKa = 10.28RR335 pKa = 11.84VPTFRR340 pKa = 11.84EE341 pKa = 3.27AALAIKK347 pKa = 9.8KK348 pKa = 10.33RR349 pKa = 11.84NLNVPDD355 pKa = 3.92LQQVFFEE362 pKa = 4.65DD363 pKa = 3.63EE364 pKa = 3.84EE365 pKa = 4.12ARR367 pKa = 11.84RR368 pKa = 11.84IANRR372 pKa = 11.84FINTVLDD379 pKa = 4.21PNKK382 pKa = 9.9LAQFPGYY389 pKa = 9.69ISEE392 pKa = 4.62GEE394 pKa = 3.3IGYY397 pKa = 9.03YY398 pKa = 10.14NKK400 pKa = 10.62YY401 pKa = 9.06LTGKK405 pKa = 10.48AIDD408 pKa = 4.01PDD410 pKa = 4.08MFVDD414 pKa = 4.38PCALVSMDD422 pKa = 4.87KK423 pKa = 9.87YY424 pKa = 10.83RR425 pKa = 11.84HH426 pKa = 5.56MIKK429 pKa = 7.8TTLKK433 pKa = 10.61PMEE436 pKa = 4.78DD437 pKa = 3.33CSGLFEE443 pKa = 5.02RR444 pKa = 11.84PLPATITYY452 pKa = 9.28HH453 pKa = 7.58DD454 pKa = 4.21KK455 pKa = 11.19GKK457 pKa = 10.97VMSTSPIFLMMANRR471 pKa = 11.84LMLCLNDD478 pKa = 4.85KK479 pKa = 10.77ISIPSGKK486 pKa = 9.13YY487 pKa = 8.54HH488 pKa = 6.33QLFSVDD494 pKa = 3.26PFAFEE499 pKa = 3.88TTKK502 pKa = 10.48EE503 pKa = 4.0FKK505 pKa = 10.61EE506 pKa = 3.73IDD508 pKa = 3.38FSKK511 pKa = 10.51FDD513 pKa = 3.39KK514 pKa = 10.68SQQRR518 pKa = 11.84LHH520 pKa = 7.41HH521 pKa = 7.01LIQFHH526 pKa = 6.82IFTALGASSEE536 pKa = 4.1FLQMWFGSHH545 pKa = 5.69EE546 pKa = 4.27LSHH549 pKa = 8.5ISDD552 pKa = 4.63GPCGIGFSVNYY563 pKa = 8.16QRR565 pKa = 11.84RR566 pKa = 11.84TGDD569 pKa = 2.69ACTYY573 pKa = 10.06LGNTIITLSALAYY586 pKa = 9.8MYY588 pKa = 10.97DD589 pKa = 3.7LLDD592 pKa = 4.26PNVTFVIASGDD603 pKa = 3.52DD604 pKa = 3.56SLIGSIKK611 pKa = 9.66PLNRR615 pKa = 11.84EE616 pKa = 3.99DD617 pKa = 3.82EE618 pKa = 4.69YY619 pKa = 11.79KK620 pKa = 9.79FTTLFNFEE628 pKa = 4.46AKK630 pKa = 9.98FPHH633 pKa = 6.26NMPFVCSKK641 pKa = 10.38FLILAPTNDD650 pKa = 3.44GGKK653 pKa = 10.0KK654 pKa = 9.37VVAVPNALKK663 pKa = 10.82LFIKK667 pKa = 10.63LGVKK671 pKa = 10.06DD672 pKa = 3.83LSPYY676 pKa = 9.92VFNEE680 pKa = 4.58WYY682 pKa = 10.58SSWLDD687 pKa = 3.86LIWYY691 pKa = 8.15FDD693 pKa = 3.65NYY695 pKa = 10.63HH696 pKa = 5.86VVSTMRR702 pKa = 11.84DD703 pKa = 3.87YY704 pKa = 11.5LSHH707 pKa = 6.83RR708 pKa = 11.84YY709 pKa = 9.32KK710 pKa = 10.82RR711 pKa = 11.84GTTFYY716 pKa = 11.03QEE718 pKa = 4.21GGMLALRR725 pKa = 11.84SVFSSKK731 pKa = 9.31TKK733 pKa = 9.91CLKK736 pKa = 10.61VLFGLEE742 pKa = 4.06PKK744 pKa = 10.41DD745 pKa = 3.64IEE747 pKa = 4.19EE748 pKa = 4.51PKK750 pKa = 9.77PKK752 pKa = 9.93KK753 pKa = 10.54VSFNQIEE760 pKa = 4.06RR761 pKa = 11.84LNFRR765 pKa = 11.84EE766 pKa = 4.53KK767 pKa = 10.74EE768 pKa = 4.06NKK770 pKa = 9.36VPNKK774 pKa = 10.12DD775 pKa = 3.05RR776 pKa = 11.84SKK778 pKa = 10.91RR779 pKa = 11.84PNKK782 pKa = 9.54SSRR785 pKa = 11.84MNSDD789 pKa = 3.31LKK791 pKa = 11.33
MM1 pKa = 7.68AFLSSMFPSDD11 pKa = 4.17SFLDD15 pKa = 4.0RR16 pKa = 11.84DD17 pKa = 3.75WEE19 pKa = 4.53VPLLLDD25 pKa = 3.5EE26 pKa = 4.74RR27 pKa = 11.84PRR29 pKa = 11.84TEE31 pKa = 3.88VEE33 pKa = 3.92QVSNEE38 pKa = 4.06EE39 pKa = 3.79VDD41 pKa = 3.78TNALFLKK48 pKa = 10.43SYY50 pKa = 10.87LLTLFLSPKK59 pKa = 9.74FKK61 pKa = 11.17YY62 pKa = 10.14IFEE65 pKa = 5.1DD66 pKa = 4.0FLGTEE71 pKa = 4.58FACYY75 pKa = 9.86VKK77 pKa = 10.34PRR79 pKa = 11.84WAIVFGCEE87 pKa = 3.14VDD89 pKa = 4.05YY90 pKa = 10.99EE91 pKa = 4.35VHH93 pKa = 6.4EE94 pKa = 4.46LVNDD98 pKa = 3.92EE99 pKa = 5.22LPLHH103 pKa = 6.88LDD105 pKa = 3.31TYY107 pKa = 9.87EE108 pKa = 3.94PWEE111 pKa = 4.34GVDD114 pKa = 6.1DD115 pKa = 4.68EE116 pKa = 4.97PCLGIDD122 pKa = 4.99DD123 pKa = 5.48LLDD126 pKa = 4.28RR127 pKa = 11.84DD128 pKa = 3.91ASPVEE133 pKa = 3.89RR134 pKa = 11.84DD135 pKa = 3.5DD136 pKa = 5.57VLFCLEE142 pKa = 4.92DD143 pKa = 3.81EE144 pKa = 4.63SDD146 pKa = 3.71GTTSLVLPTQTGIDD160 pKa = 4.17DD161 pKa = 5.36DD162 pKa = 4.66VDD164 pKa = 4.25SDD166 pKa = 3.77TSLKK170 pKa = 10.59VFEE173 pKa = 4.38STEE176 pKa = 3.92DD177 pKa = 3.78LEE179 pKa = 5.23DD180 pKa = 3.48VKK182 pKa = 10.53PTLKK186 pKa = 10.89SSDD189 pKa = 3.38LVLRR193 pKa = 11.84TINDD197 pKa = 3.25VDD199 pKa = 3.81TVIPVVEE206 pKa = 4.25KK207 pKa = 10.57FDD209 pKa = 3.91TNRR212 pKa = 11.84LHH214 pKa = 6.88LVKK217 pKa = 10.14EE218 pKa = 4.04VMMDD222 pKa = 3.5GFEE225 pKa = 4.43SLLIAHH231 pKa = 6.33EE232 pKa = 5.2RR233 pKa = 11.84ILSRR237 pKa = 11.84NDD239 pKa = 2.97WASLAMYY246 pKa = 9.12CPDD249 pKa = 4.33MEE251 pKa = 5.74IPTCGDD257 pKa = 3.35LLEE260 pKa = 5.48IPVKK264 pKa = 9.95HH265 pKa = 6.24TDD267 pKa = 2.83IEE269 pKa = 4.72FIQMAVDD276 pKa = 3.45EE277 pKa = 5.08LLPGVCDD284 pKa = 4.24LNDD287 pKa = 3.6SFFQEE292 pKa = 4.49LVEE295 pKa = 4.45TGDD298 pKa = 3.85IALEE302 pKa = 3.9LDD304 pKa = 3.15KK305 pKa = 11.57AKK307 pKa = 10.18IDD309 pKa = 3.2RR310 pKa = 11.84SVFNDD315 pKa = 2.81WSKK318 pKa = 11.44SKK320 pKa = 10.63KK321 pKa = 10.12CISSRR326 pKa = 11.84LNTGNISKK334 pKa = 10.28RR335 pKa = 11.84VPTFRR340 pKa = 11.84EE341 pKa = 3.27AALAIKK347 pKa = 9.8KK348 pKa = 10.33RR349 pKa = 11.84NLNVPDD355 pKa = 3.92LQQVFFEE362 pKa = 4.65DD363 pKa = 3.63EE364 pKa = 3.84EE365 pKa = 4.12ARR367 pKa = 11.84RR368 pKa = 11.84IANRR372 pKa = 11.84FINTVLDD379 pKa = 4.21PNKK382 pKa = 9.9LAQFPGYY389 pKa = 9.69ISEE392 pKa = 4.62GEE394 pKa = 3.3IGYY397 pKa = 9.03YY398 pKa = 10.14NKK400 pKa = 10.62YY401 pKa = 9.06LTGKK405 pKa = 10.48AIDD408 pKa = 4.01PDD410 pKa = 4.08MFVDD414 pKa = 4.38PCALVSMDD422 pKa = 4.87KK423 pKa = 9.87YY424 pKa = 10.83RR425 pKa = 11.84HH426 pKa = 5.56MIKK429 pKa = 7.8TTLKK433 pKa = 10.61PMEE436 pKa = 4.78DD437 pKa = 3.33CSGLFEE443 pKa = 5.02RR444 pKa = 11.84PLPATITYY452 pKa = 9.28HH453 pKa = 7.58DD454 pKa = 4.21KK455 pKa = 11.19GKK457 pKa = 10.97VMSTSPIFLMMANRR471 pKa = 11.84LMLCLNDD478 pKa = 4.85KK479 pKa = 10.77ISIPSGKK486 pKa = 9.13YY487 pKa = 8.54HH488 pKa = 6.33QLFSVDD494 pKa = 3.26PFAFEE499 pKa = 3.88TTKK502 pKa = 10.48EE503 pKa = 4.0FKK505 pKa = 10.61EE506 pKa = 3.73IDD508 pKa = 3.38FSKK511 pKa = 10.51FDD513 pKa = 3.39KK514 pKa = 10.68SQQRR518 pKa = 11.84LHH520 pKa = 7.41HH521 pKa = 7.01LIQFHH526 pKa = 6.82IFTALGASSEE536 pKa = 4.1FLQMWFGSHH545 pKa = 5.69EE546 pKa = 4.27LSHH549 pKa = 8.5ISDD552 pKa = 4.63GPCGIGFSVNYY563 pKa = 8.16QRR565 pKa = 11.84RR566 pKa = 11.84TGDD569 pKa = 2.69ACTYY573 pKa = 10.06LGNTIITLSALAYY586 pKa = 9.8MYY588 pKa = 10.97DD589 pKa = 3.7LLDD592 pKa = 4.26PNVTFVIASGDD603 pKa = 3.52DD604 pKa = 3.56SLIGSIKK611 pKa = 9.66PLNRR615 pKa = 11.84EE616 pKa = 3.99DD617 pKa = 3.82EE618 pKa = 4.69YY619 pKa = 11.79KK620 pKa = 9.79FTTLFNFEE628 pKa = 4.46AKK630 pKa = 9.98FPHH633 pKa = 6.26NMPFVCSKK641 pKa = 10.38FLILAPTNDD650 pKa = 3.44GGKK653 pKa = 10.0KK654 pKa = 9.37VVAVPNALKK663 pKa = 10.82LFIKK667 pKa = 10.63LGVKK671 pKa = 10.06DD672 pKa = 3.83LSPYY676 pKa = 9.92VFNEE680 pKa = 4.58WYY682 pKa = 10.58SSWLDD687 pKa = 3.86LIWYY691 pKa = 8.15FDD693 pKa = 3.65NYY695 pKa = 10.63HH696 pKa = 5.86VVSTMRR702 pKa = 11.84DD703 pKa = 3.87YY704 pKa = 11.5LSHH707 pKa = 6.83RR708 pKa = 11.84YY709 pKa = 9.32KK710 pKa = 10.82RR711 pKa = 11.84GTTFYY716 pKa = 11.03QEE718 pKa = 4.21GGMLALRR725 pKa = 11.84SVFSSKK731 pKa = 9.31TKK733 pKa = 9.91CLKK736 pKa = 10.61VLFGLEE742 pKa = 4.06PKK744 pKa = 10.41DD745 pKa = 3.64IEE747 pKa = 4.19EE748 pKa = 4.51PKK750 pKa = 9.77PKK752 pKa = 9.93KK753 pKa = 10.54VSFNQIEE760 pKa = 4.06RR761 pKa = 11.84LNFRR765 pKa = 11.84EE766 pKa = 4.53KK767 pKa = 10.74EE768 pKa = 4.06NKK770 pKa = 9.36VPNKK774 pKa = 10.12DD775 pKa = 3.05RR776 pKa = 11.84SKK778 pKa = 10.91RR779 pKa = 11.84PNKK782 pKa = 9.54SSRR785 pKa = 11.84MNSDD789 pKa = 3.31LKK791 pKa = 11.33
Molecular weight: 90.68 kDa
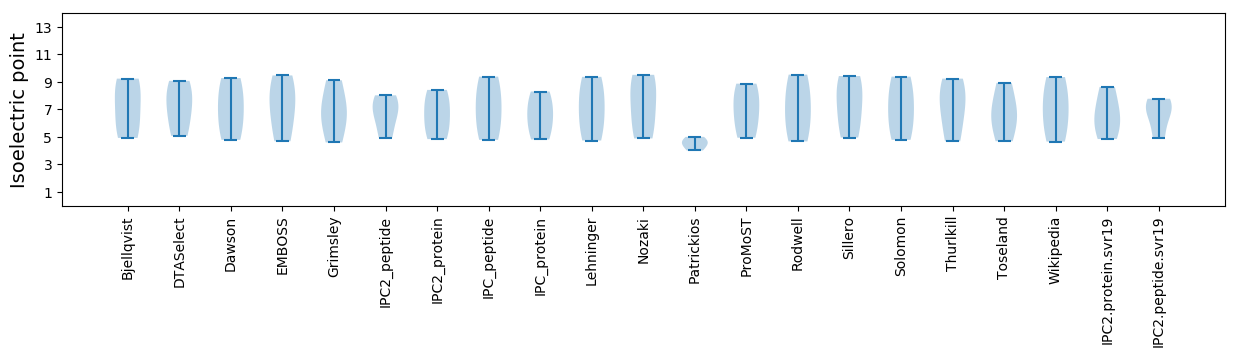
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T1WEY7|T1WEY7_9BROM ATP-dependent helicase OS=Blueberry shock virus OX=747056 PE=3 SV=1
MM1 pKa = 6.99VCKK4 pKa = 10.52FCNHH8 pKa = 4.85THH10 pKa = 7.29AGGCGQCKK18 pKa = 9.46KK19 pKa = 10.65CHH21 pKa = 4.81GTKK24 pKa = 10.41AAGPSVKK31 pKa = 10.67AQDD34 pKa = 3.36RR35 pKa = 11.84AKK37 pKa = 10.85NNPNKK42 pKa = 10.23GSPSGTKK49 pKa = 10.18SPGAKK54 pKa = 9.65GAPKK58 pKa = 10.23DD59 pKa = 3.54KK60 pKa = 10.48QVQKK64 pKa = 9.75RR65 pKa = 11.84TDD67 pKa = 3.0WTVIGPNVQPVNHH80 pKa = 7.16PNGFTLRR87 pKa = 11.84SCRR90 pKa = 11.84DD91 pKa = 3.23VQATVAGKK99 pKa = 9.64FLHH102 pKa = 6.99INFKK106 pKa = 9.4TAFPQLLNQEE116 pKa = 4.11LKK118 pKa = 9.89IYY120 pKa = 10.25SFAVRR125 pKa = 11.84CSTSIGNGWVGLVRR139 pKa = 11.84GFNPSSPTGPAVLTRR154 pKa = 11.84KK155 pKa = 10.04GFLKK159 pKa = 10.61DD160 pKa = 4.09QARR163 pKa = 11.84GWQWLAPSDD172 pKa = 3.99LEE174 pKa = 3.81YY175 pKa = 11.29DD176 pKa = 3.69KK177 pKa = 11.28FSEE180 pKa = 4.43EE181 pKa = 4.18YY182 pKa = 10.47EE183 pKa = 4.07LVFEE187 pKa = 5.03FKK189 pKa = 10.22SDD191 pKa = 3.7YY192 pKa = 10.62PIGVVMTRR200 pKa = 11.84DD201 pKa = 3.7LYY203 pKa = 11.56VVTSSLPRR211 pKa = 11.84VRR213 pKa = 11.84IPDD216 pKa = 3.72DD217 pKa = 3.7LLFVDD222 pKa = 5.08EE223 pKa = 5.82DD224 pKa = 3.89LLEE227 pKa = 4.16II228 pKa = 5.03
MM1 pKa = 6.99VCKK4 pKa = 10.52FCNHH8 pKa = 4.85THH10 pKa = 7.29AGGCGQCKK18 pKa = 9.46KK19 pKa = 10.65CHH21 pKa = 4.81GTKK24 pKa = 10.41AAGPSVKK31 pKa = 10.67AQDD34 pKa = 3.36RR35 pKa = 11.84AKK37 pKa = 10.85NNPNKK42 pKa = 10.23GSPSGTKK49 pKa = 10.18SPGAKK54 pKa = 9.65GAPKK58 pKa = 10.23DD59 pKa = 3.54KK60 pKa = 10.48QVQKK64 pKa = 9.75RR65 pKa = 11.84TDD67 pKa = 3.0WTVIGPNVQPVNHH80 pKa = 7.16PNGFTLRR87 pKa = 11.84SCRR90 pKa = 11.84DD91 pKa = 3.23VQATVAGKK99 pKa = 9.64FLHH102 pKa = 6.99INFKK106 pKa = 9.4TAFPQLLNQEE116 pKa = 4.11LKK118 pKa = 9.89IYY120 pKa = 10.25SFAVRR125 pKa = 11.84CSTSIGNGWVGLVRR139 pKa = 11.84GFNPSSPTGPAVLTRR154 pKa = 11.84KK155 pKa = 10.04GFLKK159 pKa = 10.61DD160 pKa = 4.09QARR163 pKa = 11.84GWQWLAPSDD172 pKa = 3.99LEE174 pKa = 3.81YY175 pKa = 11.29DD176 pKa = 3.69KK177 pKa = 11.28FSEE180 pKa = 4.43EE181 pKa = 4.18YY182 pKa = 10.47EE183 pKa = 4.07LVFEE187 pKa = 5.03FKK189 pKa = 10.22SDD191 pKa = 3.7YY192 pKa = 10.62PIGVVMTRR200 pKa = 11.84DD201 pKa = 3.7LYY203 pKa = 11.56VVTSSLPRR211 pKa = 11.84VRR213 pKa = 11.84IPDD216 pKa = 3.72DD217 pKa = 3.7LLFVDD222 pKa = 5.08EE223 pKa = 5.82DD224 pKa = 3.89LLEE227 pKa = 4.16II228 pKa = 5.03
Molecular weight: 25.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2359 |
228 |
1045 |
589.8 |
66.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.189 ± 0.972 | 1.95 ± 0.212 |
6.825 ± 0.783 | 6.783 ± 0.506 |
5.002 ± 0.67 | 5.256 ± 0.659 |
2.331 ± 0.234 | 5.002 ± 0.308 |
7.079 ± 0.209 | 9.496 ± 0.648 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.374 ± 0.412 | 4.324 ± 0.078 |
4.409 ± 0.811 | 2.671 ± 0.436 |
5.511 ± 0.508 | 7.418 ± 0.172 |
6.062 ± 0.386 | 7.206 ± 0.346 |
1.145 ± 0.097 | 2.967 ± 0.228 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
