
Nectarine stem pitting associated virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Luteovirus
Average proteome isoelectric point is 7.47
Get precalculated fractions of proteins
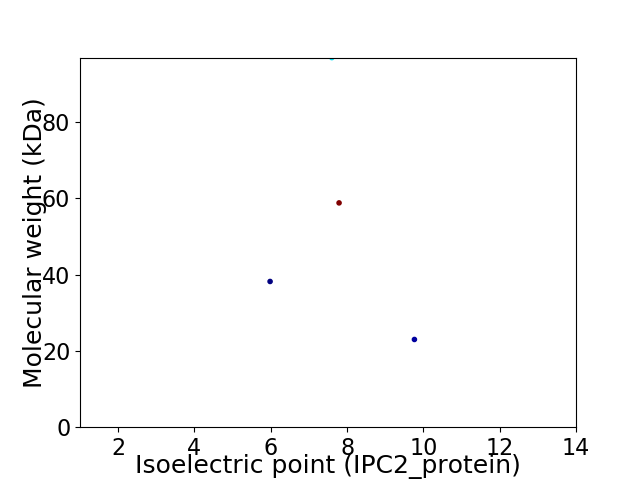
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
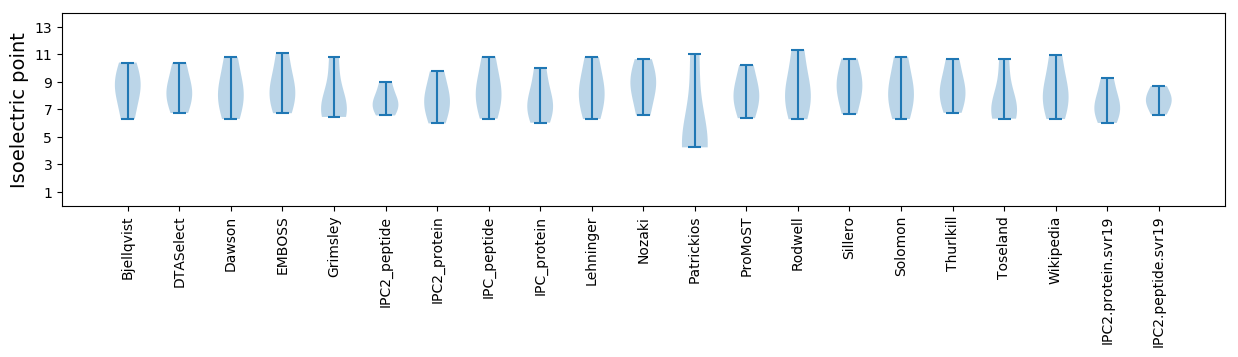
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U4HNN4|A0A0U4HNN4_9LUTE RNA dependent RNA polymerase P1 OS=Nectarine stem pitting associated virus OX=2560614 GN=NSPaV_SF04522E_gp2 PE=4 SV=1
MM1 pKa = 7.85IFDD4 pKa = 5.02LLISASTKK12 pKa = 10.14AIKK15 pKa = 10.55DD16 pKa = 4.22FISFLYY22 pKa = 10.07SKK24 pKa = 10.33CRR26 pKa = 11.84NIYY29 pKa = 10.41CRR31 pKa = 11.84FKK33 pKa = 10.41KK34 pKa = 9.71WLMDD38 pKa = 3.78FSEE41 pKa = 4.39YY42 pKa = 10.88DD43 pKa = 3.27AFVAEE48 pKa = 4.83CFEE51 pKa = 4.48TMFEE55 pKa = 4.23VEE57 pKa = 4.38TFQEE61 pKa = 4.29EE62 pKa = 4.6VVDD65 pKa = 3.8AFIRR69 pKa = 11.84IEE71 pKa = 4.39DD72 pKa = 3.65EE73 pKa = 3.86LAAAEE78 pKa = 4.25AKK80 pKa = 10.39LKK82 pKa = 10.1EE83 pKa = 4.2VHH85 pKa = 6.64NPYY88 pKa = 10.57LWGKK92 pKa = 7.6ISEE95 pKa = 4.71YY96 pKa = 10.11IFPSRR101 pKa = 11.84PEE103 pKa = 3.88DD104 pKa = 3.48VEE106 pKa = 4.33VAKK109 pKa = 10.8LQVYY113 pKa = 9.96AKK115 pKa = 8.59EE116 pKa = 3.8VQMRR120 pKa = 11.84VFIDD124 pKa = 3.48DD125 pKa = 3.91TLDD128 pKa = 3.7DD129 pKa = 3.88MEE131 pKa = 4.6EE132 pKa = 4.18AVSGTMSEE140 pKa = 4.33SQVEE144 pKa = 4.49VLALTPNQLKK154 pKa = 10.68ARR156 pKa = 11.84LKK158 pKa = 10.16KK159 pKa = 9.52AAKK162 pKa = 9.59HH163 pKa = 5.42RR164 pKa = 11.84RR165 pKa = 11.84QKK167 pKa = 10.15QAAKK171 pKa = 10.37RR172 pKa = 11.84MRR174 pKa = 11.84EE175 pKa = 3.92AMDD178 pKa = 3.39KK179 pKa = 10.56VEE181 pKa = 5.64KK182 pKa = 10.2IAEE185 pKa = 4.23LSDD188 pKa = 2.94WTTFQHH194 pKa = 6.7VEE196 pKa = 4.13VVDD199 pKa = 4.19QKK201 pKa = 10.79HH202 pKa = 4.74SHH204 pKa = 6.57PARR207 pKa = 11.84EE208 pKa = 4.16EE209 pKa = 3.66QGEE212 pKa = 4.22DD213 pKa = 3.01GKK215 pKa = 11.34KK216 pKa = 10.11IIPAKK221 pKa = 9.87IAKK224 pKa = 8.9KK225 pKa = 9.25WIWVRR230 pKa = 11.84NIKK233 pKa = 9.42TGEE236 pKa = 4.04EE237 pKa = 3.25KK238 pKa = 10.42RR239 pKa = 11.84ARR241 pKa = 11.84HH242 pKa = 6.29FIRR245 pKa = 11.84AYY247 pKa = 10.84VMSKK251 pKa = 9.69NLRR254 pKa = 11.84LRR256 pKa = 11.84GDD258 pKa = 3.81DD259 pKa = 3.52VSKK262 pKa = 10.17VTIQRR267 pKa = 11.84YY268 pKa = 8.3VEE270 pKa = 4.11QFCDD274 pKa = 3.62ANDD277 pKa = 3.77FSLEE281 pKa = 4.19AKK283 pKa = 8.29TQLIKK288 pKa = 10.61VALMMVPVPTKK299 pKa = 10.36TEE301 pKa = 3.16IDD303 pKa = 3.38MAMVVHH309 pKa = 6.77CPRR312 pKa = 11.84AEE314 pKa = 3.99ALRR317 pKa = 11.84HH318 pKa = 4.52QLEE321 pKa = 4.91CIEE324 pKa = 4.34SQVFF328 pKa = 3.22
MM1 pKa = 7.85IFDD4 pKa = 5.02LLISASTKK12 pKa = 10.14AIKK15 pKa = 10.55DD16 pKa = 4.22FISFLYY22 pKa = 10.07SKK24 pKa = 10.33CRR26 pKa = 11.84NIYY29 pKa = 10.41CRR31 pKa = 11.84FKK33 pKa = 10.41KK34 pKa = 9.71WLMDD38 pKa = 3.78FSEE41 pKa = 4.39YY42 pKa = 10.88DD43 pKa = 3.27AFVAEE48 pKa = 4.83CFEE51 pKa = 4.48TMFEE55 pKa = 4.23VEE57 pKa = 4.38TFQEE61 pKa = 4.29EE62 pKa = 4.6VVDD65 pKa = 3.8AFIRR69 pKa = 11.84IEE71 pKa = 4.39DD72 pKa = 3.65EE73 pKa = 3.86LAAAEE78 pKa = 4.25AKK80 pKa = 10.39LKK82 pKa = 10.1EE83 pKa = 4.2VHH85 pKa = 6.64NPYY88 pKa = 10.57LWGKK92 pKa = 7.6ISEE95 pKa = 4.71YY96 pKa = 10.11IFPSRR101 pKa = 11.84PEE103 pKa = 3.88DD104 pKa = 3.48VEE106 pKa = 4.33VAKK109 pKa = 10.8LQVYY113 pKa = 9.96AKK115 pKa = 8.59EE116 pKa = 3.8VQMRR120 pKa = 11.84VFIDD124 pKa = 3.48DD125 pKa = 3.91TLDD128 pKa = 3.7DD129 pKa = 3.88MEE131 pKa = 4.6EE132 pKa = 4.18AVSGTMSEE140 pKa = 4.33SQVEE144 pKa = 4.49VLALTPNQLKK154 pKa = 10.68ARR156 pKa = 11.84LKK158 pKa = 10.16KK159 pKa = 9.52AAKK162 pKa = 9.59HH163 pKa = 5.42RR164 pKa = 11.84RR165 pKa = 11.84QKK167 pKa = 10.15QAAKK171 pKa = 10.37RR172 pKa = 11.84MRR174 pKa = 11.84EE175 pKa = 3.92AMDD178 pKa = 3.39KK179 pKa = 10.56VEE181 pKa = 5.64KK182 pKa = 10.2IAEE185 pKa = 4.23LSDD188 pKa = 2.94WTTFQHH194 pKa = 6.7VEE196 pKa = 4.13VVDD199 pKa = 4.19QKK201 pKa = 10.79HH202 pKa = 4.74SHH204 pKa = 6.57PARR207 pKa = 11.84EE208 pKa = 4.16EE209 pKa = 3.66QGEE212 pKa = 4.22DD213 pKa = 3.01GKK215 pKa = 11.34KK216 pKa = 10.11IIPAKK221 pKa = 9.87IAKK224 pKa = 8.9KK225 pKa = 9.25WIWVRR230 pKa = 11.84NIKK233 pKa = 9.42TGEE236 pKa = 4.04EE237 pKa = 3.25KK238 pKa = 10.42RR239 pKa = 11.84ARR241 pKa = 11.84HH242 pKa = 6.29FIRR245 pKa = 11.84AYY247 pKa = 10.84VMSKK251 pKa = 9.69NLRR254 pKa = 11.84LRR256 pKa = 11.84GDD258 pKa = 3.81DD259 pKa = 3.52VSKK262 pKa = 10.17VTIQRR267 pKa = 11.84YY268 pKa = 8.3VEE270 pKa = 4.11QFCDD274 pKa = 3.62ANDD277 pKa = 3.77FSLEE281 pKa = 4.19AKK283 pKa = 8.29TQLIKK288 pKa = 10.61VALMMVPVPTKK299 pKa = 10.36TEE301 pKa = 3.16IDD303 pKa = 3.38MAMVVHH309 pKa = 6.77CPRR312 pKa = 11.84AEE314 pKa = 3.99ALRR317 pKa = 11.84HH318 pKa = 4.52QLEE321 pKa = 4.91CIEE324 pKa = 4.34SQVFF328 pKa = 3.22
Molecular weight: 38.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U4H028|A0A0U4H028_9LUTE Readthrough protein OS=Nectarine stem pitting associated virus OX=2560614 GN=NSPaV_SF04522E_gp3 PE=3 SV=1
MM1 pKa = 7.02SQRR4 pKa = 11.84NRR6 pKa = 11.84RR7 pKa = 11.84SQNAGWLPPMRR18 pKa = 11.84QQPPRR23 pKa = 11.84WSSQRR28 pKa = 11.84QGAPAPRR35 pKa = 11.84IVYY38 pKa = 7.96VTGPPVQPKK47 pKa = 9.66KK48 pKa = 10.12AAKK51 pKa = 9.79KK52 pKa = 9.8KK53 pKa = 10.4SPPSKK58 pKa = 10.18QPQPTSPFPAFKK70 pKa = 9.65FTIDD74 pKa = 4.22DD75 pKa = 4.15LKK77 pKa = 11.45GDD79 pKa = 3.51ASGVLKK85 pKa = 10.56FGPKK89 pKa = 9.42LDD91 pKa = 3.56QYY93 pKa = 10.94QAFVNGIMKK102 pKa = 10.28SFHH105 pKa = 7.69DD106 pKa = 3.99YY107 pKa = 10.46RR108 pKa = 11.84ISSVVIRR115 pKa = 11.84YY116 pKa = 8.41VSNAASTTPGAMAFEE131 pKa = 5.39IDD133 pKa = 3.77TSCTQTEE140 pKa = 4.29LSSKK144 pKa = 10.61VMSSPLNRR152 pKa = 11.84SFTKK156 pKa = 10.03TFSGPTIRR164 pKa = 11.84GNLWLNTKK172 pKa = 8.76QEE174 pKa = 4.3QFWLLYY180 pKa = 9.05KK181 pKa = 11.02ANGAKK186 pKa = 9.89SDD188 pKa = 3.32IAGQFIITLHH198 pKa = 5.23THH200 pKa = 4.96WQNPKK205 pKa = 9.99SS206 pKa = 3.69
MM1 pKa = 7.02SQRR4 pKa = 11.84NRR6 pKa = 11.84RR7 pKa = 11.84SQNAGWLPPMRR18 pKa = 11.84QQPPRR23 pKa = 11.84WSSQRR28 pKa = 11.84QGAPAPRR35 pKa = 11.84IVYY38 pKa = 7.96VTGPPVQPKK47 pKa = 9.66KK48 pKa = 10.12AAKK51 pKa = 9.79KK52 pKa = 9.8KK53 pKa = 10.4SPPSKK58 pKa = 10.18QPQPTSPFPAFKK70 pKa = 9.65FTIDD74 pKa = 4.22DD75 pKa = 4.15LKK77 pKa = 11.45GDD79 pKa = 3.51ASGVLKK85 pKa = 10.56FGPKK89 pKa = 9.42LDD91 pKa = 3.56QYY93 pKa = 10.94QAFVNGIMKK102 pKa = 10.28SFHH105 pKa = 7.69DD106 pKa = 3.99YY107 pKa = 10.46RR108 pKa = 11.84ISSVVIRR115 pKa = 11.84YY116 pKa = 8.41VSNAASTTPGAMAFEE131 pKa = 5.39IDD133 pKa = 3.77TSCTQTEE140 pKa = 4.29LSSKK144 pKa = 10.61VMSSPLNRR152 pKa = 11.84SFTKK156 pKa = 10.03TFSGPTIRR164 pKa = 11.84GNLWLNTKK172 pKa = 8.76QEE174 pKa = 4.3QFWLLYY180 pKa = 9.05KK181 pKa = 11.02ANGAKK186 pKa = 9.89SDD188 pKa = 3.32IAGQFIITLHH198 pKa = 5.23THH200 pKa = 4.96WQNPKK205 pKa = 9.99SS206 pKa = 3.69
Molecular weight: 23.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1907 |
206 |
847 |
476.8 |
54.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.289 ± 0.616 | 1.993 ± 0.25 |
5.349 ± 0.262 | 6.397 ± 1.14 |
5.034 ± 0.096 | 4.667 ± 0.654 |
2.307 ± 0.121 | 5.087 ± 0.445 |
7.708 ± 0.512 | 6.502 ± 0.914 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.884 ± 0.458 | 4.038 ± 0.521 |
5.611 ± 1.406 | 4.51 ± 0.592 |
5.663 ± 0.504 | 7.656 ± 0.95 |
4.772 ± 0.68 | 7.551 ± 0.456 |
1.783 ± 0.396 | 3.146 ± 0.264 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
