
Drosophila affinis sigmavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sigmavirus
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
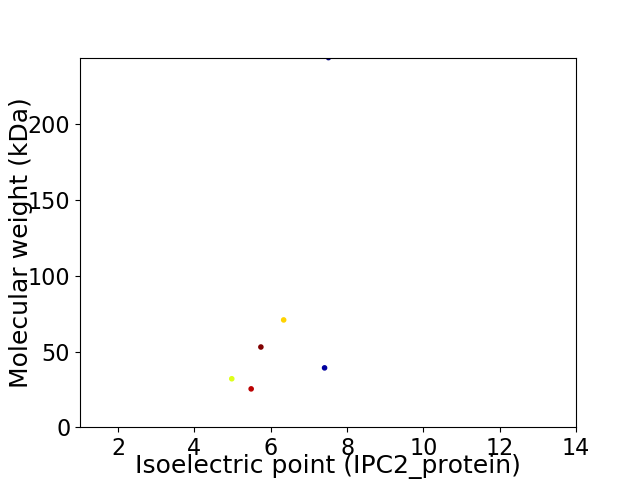
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140D8K1|A0A140D8K1_9RHAB PP3 OS=Drosophila affinis sigmavirus OX=1308859 GN=X PE=4 SV=1
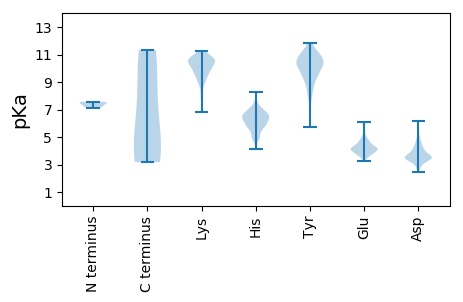
MM1 pKa = 7.56GKK3 pKa = 9.28KK4 pKa = 8.11QEE6 pKa = 4.12RR7 pKa = 11.84KK8 pKa = 9.41KK9 pKa = 11.13DD10 pKa = 3.67KK11 pKa = 11.17ADD13 pKa = 3.43TNYY16 pKa = 8.62DD17 pKa = 3.56TLVALTKK24 pKa = 10.34QAIADD29 pKa = 3.85NPGLGDD35 pKa = 3.77NLQGDD40 pKa = 4.67VNLQPASLEE49 pKa = 4.01IPGMGDD55 pKa = 3.0KK56 pKa = 11.13SEE58 pKa = 4.91LPSDD62 pKa = 4.5DD63 pKa = 4.15DD64 pKa = 5.1RR65 pKa = 11.84SDD67 pKa = 3.31GGEE70 pKa = 3.99DD71 pKa = 3.47LGDD74 pKa = 4.35IIASGGASTSQGARR88 pKa = 11.84NIIDD92 pKa = 4.07PDD94 pKa = 3.61SDD96 pKa = 3.95GEE98 pKa = 4.17SDD100 pKa = 3.89YY101 pKa = 11.77KK102 pKa = 11.09DD103 pKa = 3.75GEE105 pKa = 4.45VPLPINPKK113 pKa = 9.57RR114 pKa = 11.84VTVFYY119 pKa = 10.65RR120 pKa = 11.84PPAYY124 pKa = 9.94QHH126 pKa = 6.32AEE128 pKa = 3.81KK129 pKa = 10.15IIRR132 pKa = 11.84DD133 pKa = 3.64TVEE136 pKa = 4.0AVNKK140 pKa = 9.86LLTYY144 pKa = 10.59EE145 pKa = 4.25SVTTEE150 pKa = 3.74GHH152 pKa = 6.05GEE154 pKa = 4.03SFMLCVYY161 pKa = 10.5PRR163 pKa = 11.84MTNPVTPILEE173 pKa = 4.64NILPGHH179 pKa = 6.45RR180 pKa = 11.84EE181 pKa = 3.87HH182 pKa = 6.05VQKK185 pKa = 10.85AIEE188 pKa = 4.39STDD191 pKa = 3.4QPKK194 pKa = 10.23PCIVDD199 pKa = 3.46QPSIPIDD206 pKa = 3.37RR207 pKa = 11.84RR208 pKa = 11.84SALLEE213 pKa = 3.65KK214 pKa = 10.19NYY216 pKa = 10.11CVKK219 pKa = 10.77YY220 pKa = 10.58LDD222 pKa = 3.56QKK224 pKa = 10.31NPKK227 pKa = 9.06VYY229 pKa = 10.51KK230 pKa = 9.86IDD232 pKa = 3.52NGNPDD237 pKa = 4.07FSPEE241 pKa = 3.79KK242 pKa = 10.35FQEE245 pKa = 5.16FYY247 pKa = 10.95CKK249 pKa = 10.61YY250 pKa = 10.41QGFPKK255 pKa = 10.24NDD257 pKa = 3.35YY258 pKa = 9.96NLVRR262 pKa = 11.84RR263 pKa = 11.84MLMEE267 pKa = 3.37QSKK270 pKa = 8.7WYY272 pKa = 10.09RR273 pKa = 11.84YY274 pKa = 10.37NDD276 pKa = 3.55VLDD279 pKa = 4.38FAPLRR284 pKa = 4.14
MM1 pKa = 7.56GKK3 pKa = 9.28KK4 pKa = 8.11QEE6 pKa = 4.12RR7 pKa = 11.84KK8 pKa = 9.41KK9 pKa = 11.13DD10 pKa = 3.67KK11 pKa = 11.17ADD13 pKa = 3.43TNYY16 pKa = 8.62DD17 pKa = 3.56TLVALTKK24 pKa = 10.34QAIADD29 pKa = 3.85NPGLGDD35 pKa = 3.77NLQGDD40 pKa = 4.67VNLQPASLEE49 pKa = 4.01IPGMGDD55 pKa = 3.0KK56 pKa = 11.13SEE58 pKa = 4.91LPSDD62 pKa = 4.5DD63 pKa = 4.15DD64 pKa = 5.1RR65 pKa = 11.84SDD67 pKa = 3.31GGEE70 pKa = 3.99DD71 pKa = 3.47LGDD74 pKa = 4.35IIASGGASTSQGARR88 pKa = 11.84NIIDD92 pKa = 4.07PDD94 pKa = 3.61SDD96 pKa = 3.95GEE98 pKa = 4.17SDD100 pKa = 3.89YY101 pKa = 11.77KK102 pKa = 11.09DD103 pKa = 3.75GEE105 pKa = 4.45VPLPINPKK113 pKa = 9.57RR114 pKa = 11.84VTVFYY119 pKa = 10.65RR120 pKa = 11.84PPAYY124 pKa = 9.94QHH126 pKa = 6.32AEE128 pKa = 3.81KK129 pKa = 10.15IIRR132 pKa = 11.84DD133 pKa = 3.64TVEE136 pKa = 4.0AVNKK140 pKa = 9.86LLTYY144 pKa = 10.59EE145 pKa = 4.25SVTTEE150 pKa = 3.74GHH152 pKa = 6.05GEE154 pKa = 4.03SFMLCVYY161 pKa = 10.5PRR163 pKa = 11.84MTNPVTPILEE173 pKa = 4.64NILPGHH179 pKa = 6.45RR180 pKa = 11.84EE181 pKa = 3.87HH182 pKa = 6.05VQKK185 pKa = 10.85AIEE188 pKa = 4.39STDD191 pKa = 3.4QPKK194 pKa = 10.23PCIVDD199 pKa = 3.46QPSIPIDD206 pKa = 3.37RR207 pKa = 11.84RR208 pKa = 11.84SALLEE213 pKa = 3.65KK214 pKa = 10.19NYY216 pKa = 10.11CVKK219 pKa = 10.77YY220 pKa = 10.58LDD222 pKa = 3.56QKK224 pKa = 10.31NPKK227 pKa = 9.06VYY229 pKa = 10.51KK230 pKa = 9.86IDD232 pKa = 3.52NGNPDD237 pKa = 4.07FSPEE241 pKa = 3.79KK242 pKa = 10.35FQEE245 pKa = 5.16FYY247 pKa = 10.95CKK249 pKa = 10.61YY250 pKa = 10.41QGFPKK255 pKa = 10.24NDD257 pKa = 3.35YY258 pKa = 9.96NLVRR262 pKa = 11.84RR263 pKa = 11.84MLMEE267 pKa = 3.37QSKK270 pKa = 8.7WYY272 pKa = 10.09RR273 pKa = 11.84YY274 pKa = 10.37NDD276 pKa = 3.55VLDD279 pKa = 4.38FAPLRR284 pKa = 4.14
Molecular weight: 32.0 kDa
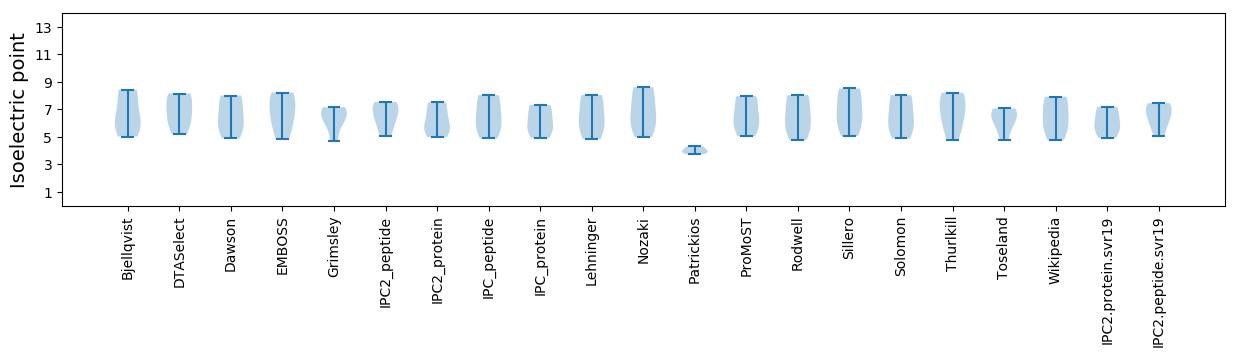
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140D8K2|A0A140D8K2_9RHAB Matrix protein OS=Drosophila affinis sigmavirus OX=1308859 GN=M PE=4 SV=1
MM1 pKa = 7.56LIIWIAQGSICSSFEE16 pKa = 4.42LICRR20 pKa = 11.84CDD22 pKa = 3.75LQNTTSLNPDD32 pKa = 2.89HH33 pKa = 6.91VARR36 pKa = 11.84AVTTYY41 pKa = 9.9YY42 pKa = 10.17IPNLDD47 pKa = 4.16RR48 pKa = 11.84YY49 pKa = 10.95NITQEE54 pKa = 3.94MKK56 pKa = 9.99IKK58 pKa = 10.28VMKK61 pKa = 9.92GGQVIDD67 pKa = 4.36TYY69 pKa = 11.61NQICLHH75 pKa = 6.42MGPMAGVTTHH85 pKa = 6.52IPVSEE90 pKa = 4.67LNSQSAAILRR100 pKa = 11.84QINSKK105 pKa = 10.61RR106 pKa = 11.84NSTVLDD112 pKa = 3.8GSVPSDD118 pKa = 3.2RR119 pKa = 11.84CRR121 pKa = 11.84RR122 pKa = 11.84GYY124 pKa = 10.78CSISTTEE131 pKa = 3.67RR132 pKa = 11.84SKK134 pKa = 11.13DD135 pKa = 3.47GKK137 pKa = 9.94PPEE140 pKa = 4.39TPVYY144 pKa = 10.16TIVTSNISIEE154 pKa = 4.02TDD156 pKa = 3.06TFNTVGKK163 pKa = 9.81EE164 pKa = 3.67VDD166 pKa = 4.72DD167 pKa = 4.74IDD169 pKa = 3.83TASYY173 pKa = 11.12LNDD176 pKa = 3.5IEE178 pKa = 5.29PMFPHH183 pKa = 6.61HH184 pKa = 7.34RR185 pKa = 11.84ILHH188 pKa = 5.85KK189 pKa = 10.69DD190 pKa = 3.27YY191 pKa = 10.97LIRR194 pKa = 11.84GLQQEE199 pKa = 4.47QEE201 pKa = 4.22TEE203 pKa = 3.72LAYY206 pKa = 10.97SKK208 pKa = 11.16NDD210 pKa = 2.99YY211 pKa = 10.36NRR213 pKa = 11.84RR214 pKa = 11.84KK215 pKa = 9.46RR216 pKa = 11.84HH217 pKa = 5.89ASPAHH222 pKa = 5.42HH223 pKa = 5.81QRR225 pKa = 11.84HH226 pKa = 5.87RR227 pKa = 11.84IINRR231 pKa = 11.84QLGMLINRR239 pKa = 11.84LNLYY243 pKa = 9.77EE244 pKa = 4.48SLWIQNYY251 pKa = 9.98IIMLVYY257 pKa = 10.3IKK259 pKa = 9.86KK260 pKa = 8.59TEE262 pKa = 4.07NPDD265 pKa = 3.06FTHH268 pKa = 6.16CVEE271 pKa = 4.25YY272 pKa = 10.09PSLRR276 pKa = 11.84DD277 pKa = 3.41MLRR280 pKa = 11.84CYY282 pKa = 11.06ADD284 pKa = 3.45NLEE287 pKa = 4.12TLYY290 pKa = 11.32NSDD293 pKa = 3.65YY294 pKa = 11.47ALDD297 pKa = 3.37AMRR300 pKa = 11.84YY301 pKa = 5.74FTEE304 pKa = 4.57GTQVKK309 pKa = 9.28YY310 pKa = 10.67NYY312 pKa = 9.8RR313 pKa = 11.84VEE315 pKa = 4.23EE316 pKa = 3.83QVKK319 pKa = 9.96YY320 pKa = 10.44KK321 pKa = 10.87LEE323 pKa = 4.04NRR325 pKa = 11.84LHH327 pKa = 5.61QKK329 pKa = 9.32RR330 pKa = 11.84HH331 pKa = 4.71NLRR334 pKa = 11.84GYY336 pKa = 10.39
MM1 pKa = 7.56LIIWIAQGSICSSFEE16 pKa = 4.42LICRR20 pKa = 11.84CDD22 pKa = 3.75LQNTTSLNPDD32 pKa = 2.89HH33 pKa = 6.91VARR36 pKa = 11.84AVTTYY41 pKa = 9.9YY42 pKa = 10.17IPNLDD47 pKa = 4.16RR48 pKa = 11.84YY49 pKa = 10.95NITQEE54 pKa = 3.94MKK56 pKa = 9.99IKK58 pKa = 10.28VMKK61 pKa = 9.92GGQVIDD67 pKa = 4.36TYY69 pKa = 11.61NQICLHH75 pKa = 6.42MGPMAGVTTHH85 pKa = 6.52IPVSEE90 pKa = 4.67LNSQSAAILRR100 pKa = 11.84QINSKK105 pKa = 10.61RR106 pKa = 11.84NSTVLDD112 pKa = 3.8GSVPSDD118 pKa = 3.2RR119 pKa = 11.84CRR121 pKa = 11.84RR122 pKa = 11.84GYY124 pKa = 10.78CSISTTEE131 pKa = 3.67RR132 pKa = 11.84SKK134 pKa = 11.13DD135 pKa = 3.47GKK137 pKa = 9.94PPEE140 pKa = 4.39TPVYY144 pKa = 10.16TIVTSNISIEE154 pKa = 4.02TDD156 pKa = 3.06TFNTVGKK163 pKa = 9.81EE164 pKa = 3.67VDD166 pKa = 4.72DD167 pKa = 4.74IDD169 pKa = 3.83TASYY173 pKa = 11.12LNDD176 pKa = 3.5IEE178 pKa = 5.29PMFPHH183 pKa = 6.61HH184 pKa = 7.34RR185 pKa = 11.84ILHH188 pKa = 5.85KK189 pKa = 10.69DD190 pKa = 3.27YY191 pKa = 10.97LIRR194 pKa = 11.84GLQQEE199 pKa = 4.47QEE201 pKa = 4.22TEE203 pKa = 3.72LAYY206 pKa = 10.97SKK208 pKa = 11.16NDD210 pKa = 2.99YY211 pKa = 10.36NRR213 pKa = 11.84RR214 pKa = 11.84KK215 pKa = 9.46RR216 pKa = 11.84HH217 pKa = 5.89ASPAHH222 pKa = 5.42HH223 pKa = 5.81QRR225 pKa = 11.84HH226 pKa = 5.87RR227 pKa = 11.84IINRR231 pKa = 11.84QLGMLINRR239 pKa = 11.84LNLYY243 pKa = 9.77EE244 pKa = 4.48SLWIQNYY251 pKa = 9.98IIMLVYY257 pKa = 10.3IKK259 pKa = 9.86KK260 pKa = 8.59TEE262 pKa = 4.07NPDD265 pKa = 3.06FTHH268 pKa = 6.16CVEE271 pKa = 4.25YY272 pKa = 10.09PSLRR276 pKa = 11.84DD277 pKa = 3.41MLRR280 pKa = 11.84CYY282 pKa = 11.06ADD284 pKa = 3.45NLEE287 pKa = 4.12TLYY290 pKa = 11.32NSDD293 pKa = 3.65YY294 pKa = 11.47ALDD297 pKa = 3.37AMRR300 pKa = 11.84YY301 pKa = 5.74FTEE304 pKa = 4.57GTQVKK309 pKa = 9.28YY310 pKa = 10.67NYY312 pKa = 9.8RR313 pKa = 11.84VEE315 pKa = 4.23EE316 pKa = 3.83QVKK319 pKa = 9.96YY320 pKa = 10.44KK321 pKa = 10.87LEE323 pKa = 4.04NRR325 pKa = 11.84LHH327 pKa = 5.61QKK329 pKa = 9.32RR330 pKa = 11.84HH331 pKa = 4.71NLRR334 pKa = 11.84GYY336 pKa = 10.39
Molecular weight: 39.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4081 |
226 |
2141 |
680.2 |
77.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.656 ± 0.636 | 1.862 ± 0.262 |
5.954 ± 0.654 | 5.146 ± 0.322 |
4.509 ± 0.584 | 5.66 ± 0.446 |
2.94 ± 0.351 | 8.037 ± 0.382 |
5.881 ± 0.374 | 8.895 ± 0.542 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.303 ± 0.167 | 5.023 ± 0.416 |
5.415 ± 0.651 | 3.48 ± 0.379 |
5.072 ± 0.501 | 7.964 ± 0.555 |
6.175 ± 0.385 | 5.391 ± 0.269 |
1.642 ± 0.265 | 3.994 ± 0.525 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
