
Ixeridium yellow mottle virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Luteoviridae; unclassified Luteoviridae
Average proteome isoelectric point is 7.4
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
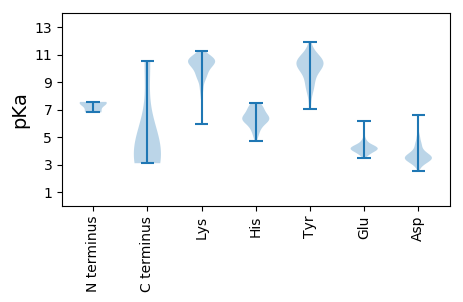
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A142D6T2|A0A142D6T2_9LUTE Serine protease OS=Ixeridium yellow mottle virus 1 OX=1809767 PE=4 SV=1
MM1 pKa = 7.56EE2 pKa = 4.32KK3 pKa = 10.53HH4 pKa = 6.21SIKK7 pKa = 10.74LFLLGLFFSSFPFFFKK23 pKa = 10.85NAFASSEE30 pKa = 4.2MEE32 pKa = 4.13LSSSLALNQSPFSNGLYY49 pKa = 10.21SPVISRR55 pKa = 11.84SLQCPISYY63 pKa = 10.5EE64 pKa = 4.23LICVEE69 pKa = 4.7KK70 pKa = 10.59PPGEE74 pKa = 4.73LISEE78 pKa = 4.53SYY80 pKa = 11.16NDD82 pKa = 5.15LIQALLEE89 pKa = 4.02KK90 pKa = 10.38SYY92 pKa = 11.89ADD94 pKa = 3.12ALSYY98 pKa = 10.03YY99 pKa = 9.86FRR101 pKa = 11.84ALEE104 pKa = 3.91TCARR108 pKa = 11.84GFATFIGYY116 pKa = 9.32SGRR119 pKa = 11.84ILSDD123 pKa = 3.78LVSMTLWFAIFLWLHH138 pKa = 5.05TFGVILSIVYY148 pKa = 10.64YY149 pKa = 11.08LMTQHH154 pKa = 6.55TVPTLILVSLTLLTRR169 pKa = 11.84YY170 pKa = 9.97IIRR173 pKa = 11.84RR174 pKa = 11.84TEE176 pKa = 3.8WVFGEE181 pKa = 4.09LPYY184 pKa = 10.92FVMRR188 pKa = 11.84ASCFLLRR195 pKa = 11.84SSYY198 pKa = 10.49QALSSSKK205 pKa = 10.61SYY207 pKa = 10.43IRR209 pKa = 11.84EE210 pKa = 3.71KK211 pKa = 10.8AVAGFTSYY219 pKa = 10.84SIPQAPPKK227 pKa = 10.67NSVLEE232 pKa = 4.09LVHH235 pKa = 7.14PDD237 pKa = 3.5GSHH240 pKa = 7.08LGYY243 pKa = 9.08ATCISLFNGEE253 pKa = 4.26NALITALHH261 pKa = 6.09CVNGNALVKK270 pKa = 10.65GKK272 pKa = 8.43TGNKK276 pKa = 9.29IPLKK280 pKa = 9.78EE281 pKa = 3.65FAARR285 pKa = 11.84IEE287 pKa = 4.45DD288 pKa = 3.72KK289 pKa = 11.28GSDD292 pKa = 3.42FALFSGPPSWEE303 pKa = 3.87SLVGCKK309 pKa = 9.11GARR312 pKa = 11.84FVTSNDD318 pKa = 3.15LARR321 pKa = 11.84CSASLYY327 pKa = 10.93KK328 pKa = 9.96LTEE331 pKa = 4.43SGWISTNAKK340 pKa = 9.6VVGPYY345 pKa = 9.47EE346 pKa = 3.89NRR348 pKa = 11.84VSVLSEE354 pKa = 3.98TFPGDD359 pKa = 2.9SGAPYY364 pKa = 9.99WNGKK368 pKa = 5.94TVLGVHH374 pKa = 6.51TGAPTHH380 pKa = 6.04EE381 pKa = 5.37NINVMTPIPHH391 pKa = 6.54IPGVTAPIYY400 pKa = 10.4VYY402 pKa = 8.85EE403 pKa = 4.34TTAPQGKK410 pKa = 9.17IFAKK414 pKa = 10.07PLNPPKK420 pKa = 10.49YY421 pKa = 10.32KK422 pKa = 10.17PIKK425 pKa = 9.41LQNHH429 pKa = 5.4WDD431 pKa = 3.65DD432 pKa = 4.46RR433 pKa = 11.84EE434 pKa = 4.38SEE436 pKa = 4.28GSSFYY441 pKa = 10.81DD442 pKa = 3.14DD443 pKa = 4.41PEE445 pKa = 3.97FLRR448 pKa = 11.84SFKK451 pKa = 9.51STKK454 pKa = 9.54SMKK457 pKa = 10.48AKK459 pKa = 10.32FIEE462 pKa = 4.42EE463 pKa = 4.45CEE465 pKa = 4.32EE466 pKa = 4.2GDD468 pKa = 3.94HH469 pKa = 7.48VISAWLYY476 pKa = 9.79SEE478 pKa = 4.39TKK480 pKa = 10.39EE481 pKa = 4.48STFLQWATTLLDD493 pKa = 4.9AILNKK498 pKa = 10.45APFEE502 pKa = 4.34HH503 pKa = 7.38DD504 pKa = 3.49VYY506 pKa = 11.61GDD508 pKa = 2.95ISTWFNTSEE517 pKa = 4.02VLTAIRR523 pKa = 11.84SYY525 pKa = 10.48EE526 pKa = 4.12ANLKK530 pKa = 8.76ATDD533 pKa = 3.47IYY535 pKa = 10.49EE536 pKa = 4.22EE537 pKa = 4.39SVSVHH542 pKa = 5.64TSDD545 pKa = 3.46TDD547 pKa = 3.92SVATAPSFVKK557 pKa = 10.59EE558 pKa = 4.09SLNRR562 pKa = 11.84ARR564 pKa = 11.84QHH566 pKa = 6.48RR567 pKa = 11.84LPTNRR572 pKa = 11.84KK573 pKa = 7.76PRR575 pKa = 11.84SKK577 pKa = 10.38HH578 pKa = 4.74RR579 pKa = 11.84RR580 pKa = 11.84HH581 pKa = 6.3PKK583 pKa = 9.95NYY585 pKa = 8.4AKK587 pKa = 10.2EE588 pKa = 3.7WRR590 pKa = 11.84RR591 pKa = 11.84NNGTDD596 pKa = 3.05PVTIISEE603 pKa = 4.19DD604 pKa = 3.27QFQGNRR610 pKa = 11.84GQDD613 pKa = 3.33CQRR616 pKa = 11.84GVKK619 pKa = 10.28EE620 pKa = 4.15SDD622 pKa = 2.84ADD624 pKa = 3.93RR625 pKa = 11.84AKK627 pKa = 10.05TRR629 pKa = 11.84RR630 pKa = 11.84EE631 pKa = 3.58RR632 pKa = 11.84TEE634 pKa = 4.23NFSQYY639 pKa = 10.5FKK641 pKa = 11.21EE642 pKa = 4.06KK643 pKa = 10.82YY644 pKa = 8.67NWEE647 pKa = 4.09TEE649 pKa = 3.99GRR651 pKa = 11.84FSDD654 pKa = 3.2IPGFRR659 pKa = 11.84DD660 pKa = 3.47CGSIPSYY667 pKa = 9.4YY668 pKa = 9.71HH669 pKa = 6.92PKK671 pKa = 10.32GKK673 pKa = 10.08QNSGWGEE680 pKa = 3.85EE681 pKa = 4.18MVANYY686 pKa = 9.61PEE688 pKa = 4.76LGEE691 pKa = 4.05AVKK694 pKa = 10.94GFGWPQFGAHH704 pKa = 6.93AEE706 pKa = 4.21LKK708 pKa = 10.44SLRR711 pKa = 11.84LQSARR716 pKa = 11.84WLEE719 pKa = 3.92RR720 pKa = 11.84LKK722 pKa = 10.81SAKK725 pKa = 9.74IPSSEE730 pKa = 3.75EE731 pKa = 3.6RR732 pKa = 11.84EE733 pKa = 4.12AVIMKK738 pKa = 8.05TLKK741 pKa = 10.26AYY743 pKa = 9.88KK744 pKa = 9.56PVEE747 pKa = 4.12TQAPDD752 pKa = 2.91IFNAKK757 pKa = 9.37EE758 pKa = 4.21DD759 pKa = 3.69ALDD762 pKa = 3.21WKK764 pKa = 10.91KK765 pKa = 10.4FLEE768 pKa = 4.3VFKK771 pKa = 11.18VAVHH775 pKa = 5.72SLEE778 pKa = 4.05LDD780 pKa = 3.19AGIGVPYY787 pKa = 9.92IAYY790 pKa = 8.45GKK792 pKa = 6.46PTHH795 pKa = 6.62RR796 pKa = 11.84GWVEE800 pKa = 3.94DD801 pKa = 4.19PEE803 pKa = 4.58LLPVLAQLTFDD814 pKa = 4.59RR815 pKa = 11.84LQKK818 pKa = 9.43MLEE821 pKa = 4.11VGYY824 pKa = 9.34EE825 pKa = 3.82AHH827 pKa = 7.27LSPEE831 pKa = 3.85QRR833 pKa = 11.84VQMGLCDD840 pKa = 5.04PIRR843 pKa = 11.84VFVKK847 pKa = 10.78GEE849 pKa = 3.59PHH851 pKa = 6.19KK852 pKa = 10.54QSKK855 pKa = 10.21LDD857 pKa = 3.38EE858 pKa = 4.27GRR860 pKa = 11.84YY861 pKa = 8.96RR862 pKa = 11.84LIMSVSLVDD871 pKa = 3.45QLVARR876 pKa = 11.84VLFQHH881 pKa = 5.11QNKK884 pKa = 9.72RR885 pKa = 11.84EE886 pKa = 3.77IDD888 pKa = 3.24LWKK891 pKa = 10.49AIPSKK896 pKa = 10.62PGFGLSTDD904 pKa = 3.65DD905 pKa = 3.46QTVEE909 pKa = 4.4FVKK912 pKa = 10.96LLAKK916 pKa = 9.84QANLQVDD923 pKa = 4.28EE924 pKa = 5.62LLQDD928 pKa = 3.88WEE930 pKa = 4.21KK931 pKa = 11.05HH932 pKa = 5.81LYY934 pKa = 7.83PTDD937 pKa = 3.45CSGFDD942 pKa = 3.27WSVSQWMLIDD952 pKa = 4.57DD953 pKa = 4.33LLVRR957 pKa = 11.84QSLQTSSFKK966 pKa = 10.66FDD968 pKa = 2.61ILQRR972 pKa = 11.84VWTDD976 pKa = 3.21CLGNSVLALSDD987 pKa = 3.49GRR989 pKa = 11.84LLAQTIPGIQKK1000 pKa = 9.54SGSYY1004 pKa = 8.29NTSSTNSRR1012 pKa = 11.84IRR1014 pKa = 11.84VMAAFHH1020 pKa = 6.85CGASWAIAMGDD1031 pKa = 3.62DD1032 pKa = 3.6ALEE1035 pKa = 4.63SKK1037 pKa = 10.96DD1038 pKa = 3.8SDD1040 pKa = 3.43LSRR1043 pKa = 11.84YY1044 pKa = 10.0AEE1046 pKa = 4.14LGFKK1050 pKa = 10.74VEE1052 pKa = 4.22VSAKK1056 pKa = 10.8LEE1058 pKa = 4.14FCSHH1062 pKa = 6.65IFEE1065 pKa = 4.48QLNLAIPVNKK1075 pKa = 10.27GKK1077 pKa = 8.79MLYY1080 pKa = 10.38KK1081 pKa = 10.66LIFGYY1086 pKa = 10.74DD1087 pKa = 3.62PGCGNQEE1094 pKa = 3.95VVKK1097 pKa = 10.62NYY1099 pKa = 10.58LDD1101 pKa = 3.66ALFSVLHH1108 pKa = 5.97EE1109 pKa = 4.48LRR1111 pKa = 11.84HH1112 pKa = 6.22DD1113 pKa = 3.91PEE1115 pKa = 4.48LVQRR1119 pKa = 11.84LHH1121 pKa = 5.86GWLVPVV1127 pKa = 4.43
MM1 pKa = 7.56EE2 pKa = 4.32KK3 pKa = 10.53HH4 pKa = 6.21SIKK7 pKa = 10.74LFLLGLFFSSFPFFFKK23 pKa = 10.85NAFASSEE30 pKa = 4.2MEE32 pKa = 4.13LSSSLALNQSPFSNGLYY49 pKa = 10.21SPVISRR55 pKa = 11.84SLQCPISYY63 pKa = 10.5EE64 pKa = 4.23LICVEE69 pKa = 4.7KK70 pKa = 10.59PPGEE74 pKa = 4.73LISEE78 pKa = 4.53SYY80 pKa = 11.16NDD82 pKa = 5.15LIQALLEE89 pKa = 4.02KK90 pKa = 10.38SYY92 pKa = 11.89ADD94 pKa = 3.12ALSYY98 pKa = 10.03YY99 pKa = 9.86FRR101 pKa = 11.84ALEE104 pKa = 3.91TCARR108 pKa = 11.84GFATFIGYY116 pKa = 9.32SGRR119 pKa = 11.84ILSDD123 pKa = 3.78LVSMTLWFAIFLWLHH138 pKa = 5.05TFGVILSIVYY148 pKa = 10.64YY149 pKa = 11.08LMTQHH154 pKa = 6.55TVPTLILVSLTLLTRR169 pKa = 11.84YY170 pKa = 9.97IIRR173 pKa = 11.84RR174 pKa = 11.84TEE176 pKa = 3.8WVFGEE181 pKa = 4.09LPYY184 pKa = 10.92FVMRR188 pKa = 11.84ASCFLLRR195 pKa = 11.84SSYY198 pKa = 10.49QALSSSKK205 pKa = 10.61SYY207 pKa = 10.43IRR209 pKa = 11.84EE210 pKa = 3.71KK211 pKa = 10.8AVAGFTSYY219 pKa = 10.84SIPQAPPKK227 pKa = 10.67NSVLEE232 pKa = 4.09LVHH235 pKa = 7.14PDD237 pKa = 3.5GSHH240 pKa = 7.08LGYY243 pKa = 9.08ATCISLFNGEE253 pKa = 4.26NALITALHH261 pKa = 6.09CVNGNALVKK270 pKa = 10.65GKK272 pKa = 8.43TGNKK276 pKa = 9.29IPLKK280 pKa = 9.78EE281 pKa = 3.65FAARR285 pKa = 11.84IEE287 pKa = 4.45DD288 pKa = 3.72KK289 pKa = 11.28GSDD292 pKa = 3.42FALFSGPPSWEE303 pKa = 3.87SLVGCKK309 pKa = 9.11GARR312 pKa = 11.84FVTSNDD318 pKa = 3.15LARR321 pKa = 11.84CSASLYY327 pKa = 10.93KK328 pKa = 9.96LTEE331 pKa = 4.43SGWISTNAKK340 pKa = 9.6VVGPYY345 pKa = 9.47EE346 pKa = 3.89NRR348 pKa = 11.84VSVLSEE354 pKa = 3.98TFPGDD359 pKa = 2.9SGAPYY364 pKa = 9.99WNGKK368 pKa = 5.94TVLGVHH374 pKa = 6.51TGAPTHH380 pKa = 6.04EE381 pKa = 5.37NINVMTPIPHH391 pKa = 6.54IPGVTAPIYY400 pKa = 10.4VYY402 pKa = 8.85EE403 pKa = 4.34TTAPQGKK410 pKa = 9.17IFAKK414 pKa = 10.07PLNPPKK420 pKa = 10.49YY421 pKa = 10.32KK422 pKa = 10.17PIKK425 pKa = 9.41LQNHH429 pKa = 5.4WDD431 pKa = 3.65DD432 pKa = 4.46RR433 pKa = 11.84EE434 pKa = 4.38SEE436 pKa = 4.28GSSFYY441 pKa = 10.81DD442 pKa = 3.14DD443 pKa = 4.41PEE445 pKa = 3.97FLRR448 pKa = 11.84SFKK451 pKa = 9.51STKK454 pKa = 9.54SMKK457 pKa = 10.48AKK459 pKa = 10.32FIEE462 pKa = 4.42EE463 pKa = 4.45CEE465 pKa = 4.32EE466 pKa = 4.2GDD468 pKa = 3.94HH469 pKa = 7.48VISAWLYY476 pKa = 9.79SEE478 pKa = 4.39TKK480 pKa = 10.39EE481 pKa = 4.48STFLQWATTLLDD493 pKa = 4.9AILNKK498 pKa = 10.45APFEE502 pKa = 4.34HH503 pKa = 7.38DD504 pKa = 3.49VYY506 pKa = 11.61GDD508 pKa = 2.95ISTWFNTSEE517 pKa = 4.02VLTAIRR523 pKa = 11.84SYY525 pKa = 10.48EE526 pKa = 4.12ANLKK530 pKa = 8.76ATDD533 pKa = 3.47IYY535 pKa = 10.49EE536 pKa = 4.22EE537 pKa = 4.39SVSVHH542 pKa = 5.64TSDD545 pKa = 3.46TDD547 pKa = 3.92SVATAPSFVKK557 pKa = 10.59EE558 pKa = 4.09SLNRR562 pKa = 11.84ARR564 pKa = 11.84QHH566 pKa = 6.48RR567 pKa = 11.84LPTNRR572 pKa = 11.84KK573 pKa = 7.76PRR575 pKa = 11.84SKK577 pKa = 10.38HH578 pKa = 4.74RR579 pKa = 11.84RR580 pKa = 11.84HH581 pKa = 6.3PKK583 pKa = 9.95NYY585 pKa = 8.4AKK587 pKa = 10.2EE588 pKa = 3.7WRR590 pKa = 11.84RR591 pKa = 11.84NNGTDD596 pKa = 3.05PVTIISEE603 pKa = 4.19DD604 pKa = 3.27QFQGNRR610 pKa = 11.84GQDD613 pKa = 3.33CQRR616 pKa = 11.84GVKK619 pKa = 10.28EE620 pKa = 4.15SDD622 pKa = 2.84ADD624 pKa = 3.93RR625 pKa = 11.84AKK627 pKa = 10.05TRR629 pKa = 11.84RR630 pKa = 11.84EE631 pKa = 3.58RR632 pKa = 11.84TEE634 pKa = 4.23NFSQYY639 pKa = 10.5FKK641 pKa = 11.21EE642 pKa = 4.06KK643 pKa = 10.82YY644 pKa = 8.67NWEE647 pKa = 4.09TEE649 pKa = 3.99GRR651 pKa = 11.84FSDD654 pKa = 3.2IPGFRR659 pKa = 11.84DD660 pKa = 3.47CGSIPSYY667 pKa = 9.4YY668 pKa = 9.71HH669 pKa = 6.92PKK671 pKa = 10.32GKK673 pKa = 10.08QNSGWGEE680 pKa = 3.85EE681 pKa = 4.18MVANYY686 pKa = 9.61PEE688 pKa = 4.76LGEE691 pKa = 4.05AVKK694 pKa = 10.94GFGWPQFGAHH704 pKa = 6.93AEE706 pKa = 4.21LKK708 pKa = 10.44SLRR711 pKa = 11.84LQSARR716 pKa = 11.84WLEE719 pKa = 3.92RR720 pKa = 11.84LKK722 pKa = 10.81SAKK725 pKa = 9.74IPSSEE730 pKa = 3.75EE731 pKa = 3.6RR732 pKa = 11.84EE733 pKa = 4.12AVIMKK738 pKa = 8.05TLKK741 pKa = 10.26AYY743 pKa = 9.88KK744 pKa = 9.56PVEE747 pKa = 4.12TQAPDD752 pKa = 2.91IFNAKK757 pKa = 9.37EE758 pKa = 4.21DD759 pKa = 3.69ALDD762 pKa = 3.21WKK764 pKa = 10.91KK765 pKa = 10.4FLEE768 pKa = 4.3VFKK771 pKa = 11.18VAVHH775 pKa = 5.72SLEE778 pKa = 4.05LDD780 pKa = 3.19AGIGVPYY787 pKa = 9.92IAYY790 pKa = 8.45GKK792 pKa = 6.46PTHH795 pKa = 6.62RR796 pKa = 11.84GWVEE800 pKa = 3.94DD801 pKa = 4.19PEE803 pKa = 4.58LLPVLAQLTFDD814 pKa = 4.59RR815 pKa = 11.84LQKK818 pKa = 9.43MLEE821 pKa = 4.11VGYY824 pKa = 9.34EE825 pKa = 3.82AHH827 pKa = 7.27LSPEE831 pKa = 3.85QRR833 pKa = 11.84VQMGLCDD840 pKa = 5.04PIRR843 pKa = 11.84VFVKK847 pKa = 10.78GEE849 pKa = 3.59PHH851 pKa = 6.19KK852 pKa = 10.54QSKK855 pKa = 10.21LDD857 pKa = 3.38EE858 pKa = 4.27GRR860 pKa = 11.84YY861 pKa = 8.96RR862 pKa = 11.84LIMSVSLVDD871 pKa = 3.45QLVARR876 pKa = 11.84VLFQHH881 pKa = 5.11QNKK884 pKa = 9.72RR885 pKa = 11.84EE886 pKa = 3.77IDD888 pKa = 3.24LWKK891 pKa = 10.49AIPSKK896 pKa = 10.62PGFGLSTDD904 pKa = 3.65DD905 pKa = 3.46QTVEE909 pKa = 4.4FVKK912 pKa = 10.96LLAKK916 pKa = 9.84QANLQVDD923 pKa = 4.28EE924 pKa = 5.62LLQDD928 pKa = 3.88WEE930 pKa = 4.21KK931 pKa = 11.05HH932 pKa = 5.81LYY934 pKa = 7.83PTDD937 pKa = 3.45CSGFDD942 pKa = 3.27WSVSQWMLIDD952 pKa = 4.57DD953 pKa = 4.33LLVRR957 pKa = 11.84QSLQTSSFKK966 pKa = 10.66FDD968 pKa = 2.61ILQRR972 pKa = 11.84VWTDD976 pKa = 3.21CLGNSVLALSDD987 pKa = 3.49GRR989 pKa = 11.84LLAQTIPGIQKK1000 pKa = 9.54SGSYY1004 pKa = 8.29NTSSTNSRR1012 pKa = 11.84IRR1014 pKa = 11.84VMAAFHH1020 pKa = 6.85CGASWAIAMGDD1031 pKa = 3.62DD1032 pKa = 3.6ALEE1035 pKa = 4.63SKK1037 pKa = 10.96DD1038 pKa = 3.8SDD1040 pKa = 3.43LSRR1043 pKa = 11.84YY1044 pKa = 10.0AEE1046 pKa = 4.14LGFKK1050 pKa = 10.74VEE1052 pKa = 4.22VSAKK1056 pKa = 10.8LEE1058 pKa = 4.14FCSHH1062 pKa = 6.65IFEE1065 pKa = 4.48QLNLAIPVNKK1075 pKa = 10.27GKK1077 pKa = 8.79MLYY1080 pKa = 10.38KK1081 pKa = 10.66LIFGYY1086 pKa = 10.74DD1087 pKa = 3.62PGCGNQEE1094 pKa = 3.95VVKK1097 pKa = 10.62NYY1099 pKa = 10.58LDD1101 pKa = 3.66ALFSVLHH1108 pKa = 5.97EE1109 pKa = 4.48LRR1111 pKa = 11.84HH1112 pKa = 6.22DD1113 pKa = 3.91PEE1115 pKa = 4.48LVQRR1119 pKa = 11.84LHH1121 pKa = 5.86GWLVPVV1127 pKa = 4.43
Molecular weight: 127.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A142D6T5|A0A142D6T5_9LUTE Movement protein OS=Ixeridium yellow mottle virus 1 OX=1809767 PE=4 SV=1
MM1 pKa = 6.79NTGAGRR7 pKa = 11.84NVGRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84TNTIPRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84NRR26 pKa = 11.84VVVVQASRR34 pKa = 11.84QPQRR38 pKa = 11.84GARR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84VRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84PAGGSGVGVRR58 pKa = 11.84RR59 pKa = 11.84SRR61 pKa = 11.84EE62 pKa = 3.88TFVFTKK68 pKa = 10.73DD69 pKa = 3.5SIAGSASGSITFGPSLSEE87 pKa = 4.04SPAFSSGILRR97 pKa = 11.84AYY99 pKa = 10.19HH100 pKa = 6.41EE101 pKa = 4.51YY102 pKa = 10.47KK103 pKa = 9.26ITMVKK108 pKa = 10.64LEE110 pKa = 4.9FISEE114 pKa = 4.18ASSTSSGSISYY125 pKa = 10.24EE126 pKa = 3.71LDD128 pKa = 3.27PHH130 pKa = 6.74CKK132 pKa = 9.93SSSLQSTVNKK142 pKa = 10.31FGITKK147 pKa = 10.34NGAKK151 pKa = 8.62TWTARR156 pKa = 11.84LINGQEE162 pKa = 3.63WHH164 pKa = 7.45DD165 pKa = 3.62ATEE168 pKa = 4.03DD169 pKa = 3.41QFRR172 pKa = 11.84ILYY175 pKa = 8.98KK176 pKa = 10.99GNGASSVAGSFRR188 pKa = 11.84ITITCQLQNPKK199 pKa = 10.55
MM1 pKa = 6.79NTGAGRR7 pKa = 11.84NVGRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84TNTIPRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84NRR26 pKa = 11.84VVVVQASRR34 pKa = 11.84QPQRR38 pKa = 11.84GARR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84VRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84PAGGSGVGVRR58 pKa = 11.84RR59 pKa = 11.84SRR61 pKa = 11.84EE62 pKa = 3.88TFVFTKK68 pKa = 10.73DD69 pKa = 3.5SIAGSASGSITFGPSLSEE87 pKa = 4.04SPAFSSGILRR97 pKa = 11.84AYY99 pKa = 10.19HH100 pKa = 6.41EE101 pKa = 4.51YY102 pKa = 10.47KK103 pKa = 9.26ITMVKK108 pKa = 10.64LEE110 pKa = 4.9FISEE114 pKa = 4.18ASSTSSGSISYY125 pKa = 10.24EE126 pKa = 3.71LDD128 pKa = 3.27PHH130 pKa = 6.74CKK132 pKa = 9.93SSSLQSTVNKK142 pKa = 10.31FGITKK147 pKa = 10.34NGAKK151 pKa = 8.62TWTARR156 pKa = 11.84LINGQEE162 pKa = 3.63WHH164 pKa = 7.45DD165 pKa = 3.62ATEE168 pKa = 4.03DD169 pKa = 3.41QFRR172 pKa = 11.84ILYY175 pKa = 8.98KK176 pKa = 10.99GNGASSVAGSFRR188 pKa = 11.84ITITCQLQNPKK199 pKa = 10.55
Molecular weight: 22.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3125 |
183 |
1127 |
520.8 |
58.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.688 ± 0.248 | 1.408 ± 0.121 |
4.288 ± 0.434 | 6.528 ± 0.28 |
4.448 ± 0.5 | 6.528 ± 0.37 |
2.176 ± 0.215 | 4.928 ± 0.341 |
5.44 ± 0.622 | 8.64 ± 1.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.472 ± 0.112 | 4.16 ± 0.358 |
6.24 ± 0.674 | 3.552 ± 0.51 |
6.464 ± 1.098 | 10.112 ± 0.699 |
6.048 ± 0.608 | 5.568 ± 0.245 |
1.888 ± 0.159 | 3.392 ± 0.298 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
