
Duck faeces associated circular DNA virus 2
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.84
Get precalculated fractions of proteins
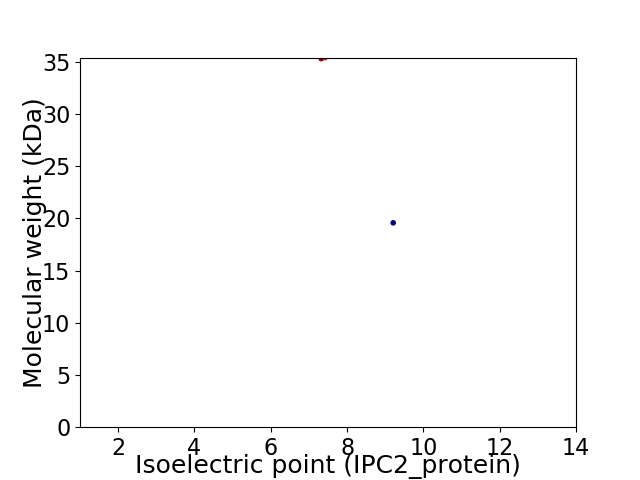
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
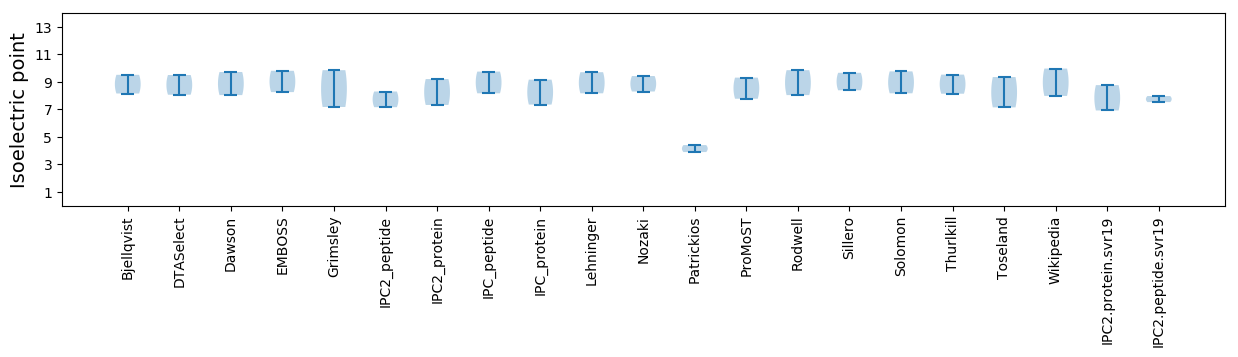
Protein with the lowest isoelectric point:
>tr|A0A160HX33|A0A160HX33_9VIRU Replication associated protein OS=Duck faeces associated circular DNA virus 2 OX=1843769 PE=3 SV=1
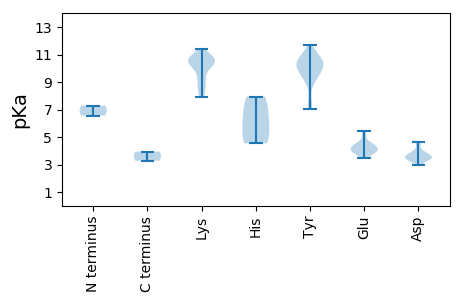
MM1 pKa = 6.56TTPIAEE7 pKa = 4.51DD8 pKa = 3.42NDD10 pKa = 3.83FKK12 pKa = 11.37FQGIRR17 pKa = 11.84ALLTYY22 pKa = 7.05PQCNEE27 pKa = 3.49GSIEE31 pKa = 3.94EE32 pKa = 4.57LLTFFKK38 pKa = 10.62TIFGNKK44 pKa = 7.91IEE46 pKa = 4.56YY47 pKa = 10.07IVVCKK52 pKa = 10.21EE53 pKa = 3.69NHH55 pKa = 6.35HH56 pKa = 5.74EE57 pKa = 4.13TDD59 pKa = 3.44GEE61 pKa = 4.45HH62 pKa = 4.54YY63 pKa = 10.26HH64 pKa = 7.94AYY66 pKa = 9.82IKK68 pKa = 10.77FNTRR72 pKa = 11.84TRR74 pKa = 11.84VYY76 pKa = 11.07ANNLVYY82 pKa = 10.48RR83 pKa = 11.84GVRR86 pKa = 11.84PNIEE90 pKa = 4.02KK91 pKa = 8.83VTRR94 pKa = 11.84TAYY97 pKa = 10.5AAVNYY102 pKa = 9.3VKK104 pKa = 10.66KK105 pKa = 10.68DD106 pKa = 3.51GNFKK110 pKa = 11.11EE111 pKa = 5.13EE112 pKa = 4.52GTCTDD117 pKa = 2.95IKK119 pKa = 11.08KK120 pKa = 8.77LTTQEE125 pKa = 3.78KK126 pKa = 10.0YY127 pKa = 10.97KK128 pKa = 10.67LIKK131 pKa = 8.46EE132 pKa = 4.12KK133 pKa = 10.59KK134 pKa = 7.93YY135 pKa = 10.73IEE137 pKa = 4.24IFEE140 pKa = 4.41MATLSLPEE148 pKa = 4.11LCKK151 pKa = 10.6VKK153 pKa = 10.45QIQRR157 pKa = 11.84EE158 pKa = 4.21LIINDD163 pKa = 3.41WPLDD167 pKa = 3.45GSKK170 pKa = 10.34NRR172 pKa = 11.84KK173 pKa = 9.28VYY175 pKa = 9.31WFYY178 pKa = 11.73GKK180 pKa = 8.6TGTGKK185 pKa = 8.1TRR187 pKa = 11.84EE188 pKa = 4.23ATKK191 pKa = 10.61MMIEE195 pKa = 4.62LYY197 pKa = 10.25GEE199 pKa = 3.88NWISLTGDD207 pKa = 3.03LRR209 pKa = 11.84TFFDD213 pKa = 4.61PYY215 pKa = 10.64NGEE218 pKa = 3.97KK219 pKa = 10.68GVIFDD224 pKa = 4.59DD225 pKa = 3.72VRR227 pKa = 11.84KK228 pKa = 10.38GSIIWNTLLTITDD241 pKa = 4.2RR242 pKa = 11.84YY243 pKa = 7.67RR244 pKa = 11.84TSVNVKK250 pKa = 9.63GSRR253 pKa = 11.84IPWLAEE259 pKa = 3.81TIIFTSPQHH268 pKa = 4.68FTEE271 pKa = 4.38VFTTEE276 pKa = 3.84KK277 pKa = 10.66DD278 pKa = 3.35GEE280 pKa = 4.16RR281 pKa = 11.84AQWDD285 pKa = 3.68GLEE288 pKa = 3.96QFEE291 pKa = 5.08RR292 pKa = 11.84RR293 pKa = 11.84ISEE296 pKa = 3.63IKK298 pKa = 10.33EE299 pKa = 3.88FNN301 pKa = 3.27
MM1 pKa = 6.56TTPIAEE7 pKa = 4.51DD8 pKa = 3.42NDD10 pKa = 3.83FKK12 pKa = 11.37FQGIRR17 pKa = 11.84ALLTYY22 pKa = 7.05PQCNEE27 pKa = 3.49GSIEE31 pKa = 3.94EE32 pKa = 4.57LLTFFKK38 pKa = 10.62TIFGNKK44 pKa = 7.91IEE46 pKa = 4.56YY47 pKa = 10.07IVVCKK52 pKa = 10.21EE53 pKa = 3.69NHH55 pKa = 6.35HH56 pKa = 5.74EE57 pKa = 4.13TDD59 pKa = 3.44GEE61 pKa = 4.45HH62 pKa = 4.54YY63 pKa = 10.26HH64 pKa = 7.94AYY66 pKa = 9.82IKK68 pKa = 10.77FNTRR72 pKa = 11.84TRR74 pKa = 11.84VYY76 pKa = 11.07ANNLVYY82 pKa = 10.48RR83 pKa = 11.84GVRR86 pKa = 11.84PNIEE90 pKa = 4.02KK91 pKa = 8.83VTRR94 pKa = 11.84TAYY97 pKa = 10.5AAVNYY102 pKa = 9.3VKK104 pKa = 10.66KK105 pKa = 10.68DD106 pKa = 3.51GNFKK110 pKa = 11.11EE111 pKa = 5.13EE112 pKa = 4.52GTCTDD117 pKa = 2.95IKK119 pKa = 11.08KK120 pKa = 8.77LTTQEE125 pKa = 3.78KK126 pKa = 10.0YY127 pKa = 10.97KK128 pKa = 10.67LIKK131 pKa = 8.46EE132 pKa = 4.12KK133 pKa = 10.59KK134 pKa = 7.93YY135 pKa = 10.73IEE137 pKa = 4.24IFEE140 pKa = 4.41MATLSLPEE148 pKa = 4.11LCKK151 pKa = 10.6VKK153 pKa = 10.45QIQRR157 pKa = 11.84EE158 pKa = 4.21LIINDD163 pKa = 3.41WPLDD167 pKa = 3.45GSKK170 pKa = 10.34NRR172 pKa = 11.84KK173 pKa = 9.28VYY175 pKa = 9.31WFYY178 pKa = 11.73GKK180 pKa = 8.6TGTGKK185 pKa = 8.1TRR187 pKa = 11.84EE188 pKa = 4.23ATKK191 pKa = 10.61MMIEE195 pKa = 4.62LYY197 pKa = 10.25GEE199 pKa = 3.88NWISLTGDD207 pKa = 3.03LRR209 pKa = 11.84TFFDD213 pKa = 4.61PYY215 pKa = 10.64NGEE218 pKa = 3.97KK219 pKa = 10.68GVIFDD224 pKa = 4.59DD225 pKa = 3.72VRR227 pKa = 11.84KK228 pKa = 10.38GSIIWNTLLTITDD241 pKa = 4.2RR242 pKa = 11.84YY243 pKa = 7.67RR244 pKa = 11.84TSVNVKK250 pKa = 9.63GSRR253 pKa = 11.84IPWLAEE259 pKa = 3.81TIIFTSPQHH268 pKa = 4.68FTEE271 pKa = 4.38VFTTEE276 pKa = 3.84KK277 pKa = 10.66DD278 pKa = 3.35GEE280 pKa = 4.16RR281 pKa = 11.84AQWDD285 pKa = 3.68GLEE288 pKa = 3.96QFEE291 pKa = 5.08RR292 pKa = 11.84RR293 pKa = 11.84ISEE296 pKa = 3.63IKK298 pKa = 10.33EE299 pKa = 3.88FNN301 pKa = 3.27
Molecular weight: 35.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A160HX33|A0A160HX33_9VIRU Replication associated protein OS=Duck faeces associated circular DNA virus 2 OX=1843769 PE=3 SV=1
MM1 pKa = 7.28PRR3 pKa = 11.84YY4 pKa = 9.47YY5 pKa = 10.39KK6 pKa = 10.29YY7 pKa = 10.12KK8 pKa = 8.93RR9 pKa = 11.84YY10 pKa = 8.87YY11 pKa = 10.19RR12 pKa = 11.84RR13 pKa = 11.84MYY15 pKa = 9.38PKK17 pKa = 10.21KK18 pKa = 9.42RR19 pKa = 11.84WASNIFTEE27 pKa = 4.22NNIVTVLASQKK38 pKa = 10.7DD39 pKa = 3.79AFASTTICANSPQTDD54 pKa = 3.48VPTPVIVKK62 pKa = 9.31FGRR65 pKa = 11.84CKK67 pKa = 10.4VKK69 pKa = 10.74GDD71 pKa = 3.45VRR73 pKa = 11.84TDD75 pKa = 3.33LDD77 pKa = 3.5NGNNFVSATMYY88 pKa = 10.31LAYY91 pKa = 10.52VPEE94 pKa = 5.45GYY96 pKa = 9.73PIADD100 pKa = 3.91LPKK103 pKa = 10.11LHH105 pKa = 7.36PEE107 pKa = 4.23YY108 pKa = 10.47IIGWTQISLDD118 pKa = 3.74SGNTFSFSSPLKK130 pKa = 10.41RR131 pKa = 11.84NLNSGDD137 pKa = 3.7KK138 pKa = 10.69IMMLFYY144 pKa = 11.01VSATNTAQYY153 pKa = 10.56VRR155 pKa = 11.84AFNFYY160 pKa = 9.53YY161 pKa = 9.8TVQYY165 pKa = 7.15WTTSAA170 pKa = 3.94
MM1 pKa = 7.28PRR3 pKa = 11.84YY4 pKa = 9.47YY5 pKa = 10.39KK6 pKa = 10.29YY7 pKa = 10.12KK8 pKa = 8.93RR9 pKa = 11.84YY10 pKa = 8.87YY11 pKa = 10.19RR12 pKa = 11.84RR13 pKa = 11.84MYY15 pKa = 9.38PKK17 pKa = 10.21KK18 pKa = 9.42RR19 pKa = 11.84WASNIFTEE27 pKa = 4.22NNIVTVLASQKK38 pKa = 10.7DD39 pKa = 3.79AFASTTICANSPQTDD54 pKa = 3.48VPTPVIVKK62 pKa = 9.31FGRR65 pKa = 11.84CKK67 pKa = 10.4VKK69 pKa = 10.74GDD71 pKa = 3.45VRR73 pKa = 11.84TDD75 pKa = 3.33LDD77 pKa = 3.5NGNNFVSATMYY88 pKa = 10.31LAYY91 pKa = 10.52VPEE94 pKa = 5.45GYY96 pKa = 9.73PIADD100 pKa = 3.91LPKK103 pKa = 10.11LHH105 pKa = 7.36PEE107 pKa = 4.23YY108 pKa = 10.47IIGWTQISLDD118 pKa = 3.74SGNTFSFSSPLKK130 pKa = 10.41RR131 pKa = 11.84NLNSGDD137 pKa = 3.7KK138 pKa = 10.69IMMLFYY144 pKa = 11.01VSATNTAQYY153 pKa = 10.56VRR155 pKa = 11.84AFNFYY160 pKa = 9.53YY161 pKa = 9.8TVQYY165 pKa = 7.15WTTSAA170 pKa = 3.94
Molecular weight: 19.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
471 |
170 |
301 |
235.5 |
27.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.883 ± 1.203 | 1.274 ± 0.054 |
4.671 ± 0.019 | 6.794 ± 2.78 |
5.52 ± 0.125 | 5.52 ± 0.775 |
1.274 ± 0.379 | 7.431 ± 1.181 |
8.28 ± 1.0 | 5.945 ± 0.36 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.911 ± 0.57 | 6.157 ± 0.499 |
3.822 ± 1.139 | 2.76 ± 0.1 |
5.52 ± 0.125 | 4.671 ± 1.645 |
9.554 ± 0.404 | 5.732 ± 0.733 |
1.911 ± 0.081 | 6.369 ± 1.357 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
