
Streptomyces zhaozhouensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
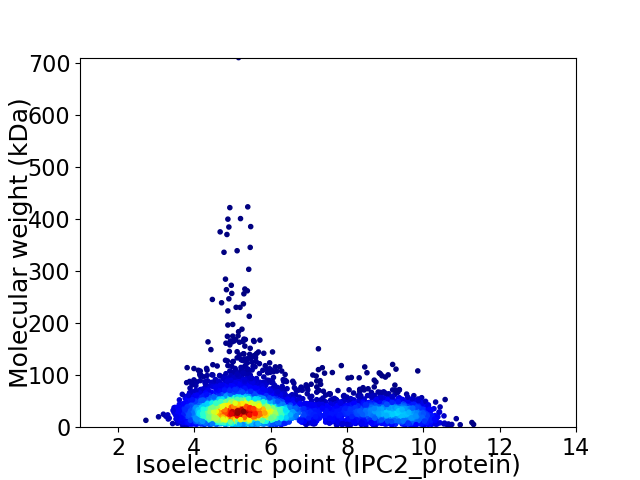
Virtual 2D-PAGE plot for 5537 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A286DWY6|A0A286DWY6_9ACTN Iron-containing redox enzyme OS=Streptomyces zhaozhouensis OX=1300267 GN=SAMN06297387_10987 PE=4 SV=1
MM1 pKa = 7.45RR2 pKa = 11.84TTGRR6 pKa = 11.84LAVSLLTATALLTAGCSSDD25 pKa = 3.82DD26 pKa = 3.74DD27 pKa = 4.31RR28 pKa = 11.84PEE30 pKa = 3.66EE31 pKa = 4.02RR32 pKa = 11.84APDD35 pKa = 3.78ADD37 pKa = 3.9ASDD40 pKa = 4.36GSSTTEE46 pKa = 4.24PPDD49 pKa = 3.94APPLEE54 pKa = 4.58PLSEE58 pKa = 4.15EE59 pKa = 4.29TPEE62 pKa = 4.0EE63 pKa = 3.83LLPYY67 pKa = 10.25YY68 pKa = 9.19EE69 pKa = 4.64QQLNWRR75 pKa = 11.84GCGAAEE81 pKa = 4.05FQCASLTVPLDD92 pKa = 3.71YY93 pKa = 10.92EE94 pKa = 4.72DD95 pKa = 5.54PNPADD100 pKa = 5.54DD101 pKa = 3.84IQLTVTRR108 pKa = 11.84SRR110 pKa = 11.84ATDD113 pKa = 3.09QDD115 pKa = 3.49EE116 pKa = 4.87RR117 pKa = 11.84IGALLMNPGGPGASAVDD134 pKa = 4.38FAQGSAEE141 pKa = 4.58FIYY144 pKa = 10.73ADD146 pKa = 3.85EE147 pKa = 4.11VRR149 pKa = 11.84ARR151 pKa = 11.84YY152 pKa = 10.61DD153 pKa = 3.29MVGLDD158 pKa = 4.25ARR160 pKa = 11.84GTGQSEE166 pKa = 4.03PVSCLEE172 pKa = 3.95GPEE175 pKa = 3.72MDD177 pKa = 5.1AYY179 pKa = 9.85TLVDD183 pKa = 3.73RR184 pKa = 11.84TPDD187 pKa = 3.42DD188 pKa = 3.71EE189 pKa = 6.55AEE191 pKa = 4.08VDD193 pKa = 3.91ALTAAQEE200 pKa = 4.03EE201 pKa = 4.82FARR204 pKa = 11.84GCQEE208 pKa = 3.47GSGALLEE215 pKa = 4.99HH216 pKa = 7.08ISTIEE221 pKa = 3.9SARR224 pKa = 11.84DD225 pKa = 3.31MDD227 pKa = 4.51VLRR230 pKa = 11.84QALGDD235 pKa = 3.76EE236 pKa = 4.18RR237 pKa = 11.84LHH239 pKa = 6.02YY240 pKa = 10.99VGFSYY245 pKa = 8.91GTKK248 pKa = 10.3LGAVYY253 pKa = 10.51AGLYY257 pKa = 6.36PQRR260 pKa = 11.84VGHH263 pKa = 6.66LVLDD267 pKa = 4.13AAMDD271 pKa = 3.93PRR273 pKa = 11.84LDD275 pKa = 3.56TLSTDD280 pKa = 3.51RR281 pKa = 11.84EE282 pKa = 4.09QAGGFEE288 pKa = 4.12TAFRR292 pKa = 11.84AFAEE296 pKa = 4.49DD297 pKa = 4.13CVSGADD303 pKa = 3.71CPLGQEE309 pKa = 4.38SADD312 pKa = 3.85DD313 pKa = 3.73ASQRR317 pKa = 11.84LLDD320 pKa = 4.43FFAAVDD326 pKa = 3.99AEE328 pKa = 4.29PLPSGDD334 pKa = 3.27EE335 pKa = 4.23RR336 pKa = 11.84EE337 pKa = 4.41LTEE340 pKa = 4.33SLATTGVAYY349 pKa = 10.3ALYY352 pKa = 10.67SEE354 pKa = 5.52DD355 pKa = 3.08LWPTLRR361 pKa = 11.84RR362 pKa = 11.84ALAAAIEE369 pKa = 4.57DD370 pKa = 4.17GQGGEE375 pKa = 4.86LLTLADD381 pKa = 4.49SYY383 pKa = 11.8NGRR386 pKa = 11.84TPGGGYY392 pKa = 8.49DD393 pKa = 3.27TDD395 pKa = 3.62MFAFPAISCLDD406 pKa = 3.77SPAGNADD413 pKa = 3.12ADD415 pKa = 3.99AVRR418 pKa = 11.84EE419 pKa = 3.87QLPSYY424 pKa = 10.4EE425 pKa = 4.22EE426 pKa = 4.03ASPTFGADD434 pKa = 3.48FAWATLVCAAWPVEE448 pKa = 4.06PSGGPVSIAAEE459 pKa = 4.23GADD462 pKa = 3.99PILVVGTTRR471 pKa = 11.84DD472 pKa = 3.33PATPYY477 pKa = 10.59AWSEE481 pKa = 3.94GLADD485 pKa = 3.77QLSSGVLLTYY495 pKa = 10.59EE496 pKa = 4.72GDD498 pKa = 2.97GHH500 pKa = 5.3TAYY503 pKa = 10.37GGLSEE508 pKa = 5.6CVDD511 pKa = 3.69STVNDD516 pKa = 3.45YY517 pKa = 11.59LLANTVPDD525 pKa = 5.55DD526 pKa = 3.67GTTCC530 pKa = 4.01
MM1 pKa = 7.45RR2 pKa = 11.84TTGRR6 pKa = 11.84LAVSLLTATALLTAGCSSDD25 pKa = 3.82DD26 pKa = 3.74DD27 pKa = 4.31RR28 pKa = 11.84PEE30 pKa = 3.66EE31 pKa = 4.02RR32 pKa = 11.84APDD35 pKa = 3.78ADD37 pKa = 3.9ASDD40 pKa = 4.36GSSTTEE46 pKa = 4.24PPDD49 pKa = 3.94APPLEE54 pKa = 4.58PLSEE58 pKa = 4.15EE59 pKa = 4.29TPEE62 pKa = 4.0EE63 pKa = 3.83LLPYY67 pKa = 10.25YY68 pKa = 9.19EE69 pKa = 4.64QQLNWRR75 pKa = 11.84GCGAAEE81 pKa = 4.05FQCASLTVPLDD92 pKa = 3.71YY93 pKa = 10.92EE94 pKa = 4.72DD95 pKa = 5.54PNPADD100 pKa = 5.54DD101 pKa = 3.84IQLTVTRR108 pKa = 11.84SRR110 pKa = 11.84ATDD113 pKa = 3.09QDD115 pKa = 3.49EE116 pKa = 4.87RR117 pKa = 11.84IGALLMNPGGPGASAVDD134 pKa = 4.38FAQGSAEE141 pKa = 4.58FIYY144 pKa = 10.73ADD146 pKa = 3.85EE147 pKa = 4.11VRR149 pKa = 11.84ARR151 pKa = 11.84YY152 pKa = 10.61DD153 pKa = 3.29MVGLDD158 pKa = 4.25ARR160 pKa = 11.84GTGQSEE166 pKa = 4.03PVSCLEE172 pKa = 3.95GPEE175 pKa = 3.72MDD177 pKa = 5.1AYY179 pKa = 9.85TLVDD183 pKa = 3.73RR184 pKa = 11.84TPDD187 pKa = 3.42DD188 pKa = 3.71EE189 pKa = 6.55AEE191 pKa = 4.08VDD193 pKa = 3.91ALTAAQEE200 pKa = 4.03EE201 pKa = 4.82FARR204 pKa = 11.84GCQEE208 pKa = 3.47GSGALLEE215 pKa = 4.99HH216 pKa = 7.08ISTIEE221 pKa = 3.9SARR224 pKa = 11.84DD225 pKa = 3.31MDD227 pKa = 4.51VLRR230 pKa = 11.84QALGDD235 pKa = 3.76EE236 pKa = 4.18RR237 pKa = 11.84LHH239 pKa = 6.02YY240 pKa = 10.99VGFSYY245 pKa = 8.91GTKK248 pKa = 10.3LGAVYY253 pKa = 10.51AGLYY257 pKa = 6.36PQRR260 pKa = 11.84VGHH263 pKa = 6.66LVLDD267 pKa = 4.13AAMDD271 pKa = 3.93PRR273 pKa = 11.84LDD275 pKa = 3.56TLSTDD280 pKa = 3.51RR281 pKa = 11.84EE282 pKa = 4.09QAGGFEE288 pKa = 4.12TAFRR292 pKa = 11.84AFAEE296 pKa = 4.49DD297 pKa = 4.13CVSGADD303 pKa = 3.71CPLGQEE309 pKa = 4.38SADD312 pKa = 3.85DD313 pKa = 3.73ASQRR317 pKa = 11.84LLDD320 pKa = 4.43FFAAVDD326 pKa = 3.99AEE328 pKa = 4.29PLPSGDD334 pKa = 3.27EE335 pKa = 4.23RR336 pKa = 11.84EE337 pKa = 4.41LTEE340 pKa = 4.33SLATTGVAYY349 pKa = 10.3ALYY352 pKa = 10.67SEE354 pKa = 5.52DD355 pKa = 3.08LWPTLRR361 pKa = 11.84RR362 pKa = 11.84ALAAAIEE369 pKa = 4.57DD370 pKa = 4.17GQGGEE375 pKa = 4.86LLTLADD381 pKa = 4.49SYY383 pKa = 11.8NGRR386 pKa = 11.84TPGGGYY392 pKa = 8.49DD393 pKa = 3.27TDD395 pKa = 3.62MFAFPAISCLDD406 pKa = 3.77SPAGNADD413 pKa = 3.12ADD415 pKa = 3.99AVRR418 pKa = 11.84EE419 pKa = 3.87QLPSYY424 pKa = 10.4EE425 pKa = 4.22EE426 pKa = 4.03ASPTFGADD434 pKa = 3.48FAWATLVCAAWPVEE448 pKa = 4.06PSGGPVSIAAEE459 pKa = 4.23GADD462 pKa = 3.99PILVVGTTRR471 pKa = 11.84DD472 pKa = 3.33PATPYY477 pKa = 10.59AWSEE481 pKa = 3.94GLADD485 pKa = 3.77QLSSGVLLTYY495 pKa = 10.59EE496 pKa = 4.72GDD498 pKa = 2.97GHH500 pKa = 5.3TAYY503 pKa = 10.37GGLSEE508 pKa = 5.6CVDD511 pKa = 3.69STVNDD516 pKa = 3.45YY517 pKa = 11.59LLANTVPDD525 pKa = 5.55DD526 pKa = 3.67GTTCC530 pKa = 4.01
Molecular weight: 56.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A286DYQ6|A0A286DYQ6_9ACTN Phosphotransferase enzyme family protein OS=Streptomyces zhaozhouensis OX=1300267 GN=SAMN06297387_112144 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIMANRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.37GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIMANRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.37GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1918315 |
28 |
6744 |
346.5 |
37.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.056 ± 0.051 | 0.72 ± 0.009 |
5.995 ± 0.03 | 6.6 ± 0.03 |
2.623 ± 0.017 | 9.787 ± 0.032 |
2.259 ± 0.016 | 2.607 ± 0.021 |
1.281 ± 0.018 | 10.764 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.573 ± 0.013 | 1.595 ± 0.016 |
6.468 ± 0.028 | 2.445 ± 0.02 |
8.794 ± 0.038 | 4.75 ± 0.021 |
5.79 ± 0.026 | 8.476 ± 0.029 |
1.54 ± 0.012 | 1.877 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
