
Clostridium sp. MSTE9
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; unclassified Clostridium
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
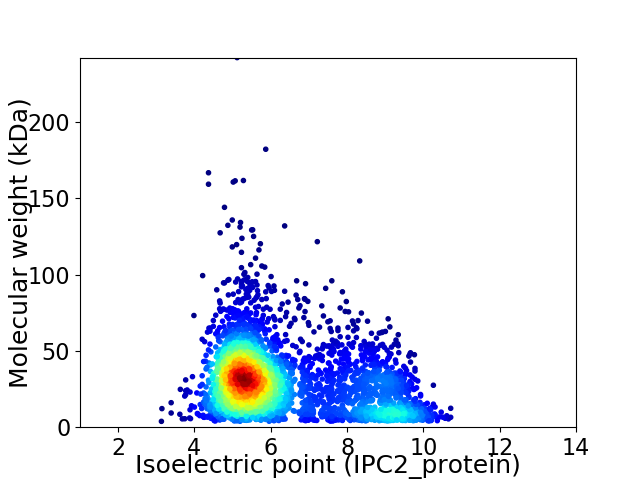
Virtual 2D-PAGE plot for 3478 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J0N6R8|J0N6R8_9CLOT Efflux transporter RND family MFP subunit OS=Clostridium sp. MSTE9 OX=1105031 GN=HMPREF1141_3061 PE=3 SV=1
MM1 pKa = 8.26DD2 pKa = 4.53ISQINSYY9 pKa = 10.88SSASTKK15 pKa = 10.23RR16 pKa = 11.84SSANSSSSLSIEE28 pKa = 4.01QFLSLLAAQLSNQDD42 pKa = 3.67VLNPTQDD49 pKa = 3.52TEE51 pKa = 5.07FVAQMAQFTSLQALEE66 pKa = 4.79NLNQYY71 pKa = 11.06ASYY74 pKa = 10.74QYY76 pKa = 11.0GSSLIGKK83 pKa = 8.42KK84 pKa = 10.48VSVAMYY90 pKa = 10.08DD91 pKa = 3.31STGKK95 pKa = 10.21YY96 pKa = 8.96VSDD99 pKa = 3.51MGVVTQADD107 pKa = 4.33YY108 pKa = 11.61SSGDD112 pKa = 3.2TTVTVNGRR120 pKa = 11.84SYY122 pKa = 11.34GIANIMEE129 pKa = 4.38VFNDD133 pKa = 3.49ASYY136 pKa = 11.6GSYY139 pKa = 10.34QYY141 pKa = 11.32AASLVGKK148 pKa = 9.37KK149 pKa = 8.76VQVTGYY155 pKa = 9.86DD156 pKa = 3.28VNGNVVEE163 pKa = 4.16NTGIVSSCTFASGEE177 pKa = 3.99AMVVVDD183 pKa = 4.02GQKK186 pKa = 8.83YY187 pKa = 6.98TLSAVRR193 pKa = 11.84QVLSQNADD201 pKa = 3.6PEE203 pKa = 4.38PTDD206 pKa = 4.51PEE208 pKa = 4.42TSDD211 pKa = 3.54DD212 pKa = 3.84TTVPDD217 pKa = 4.03TDD219 pKa = 4.0SSGSTDD225 pKa = 3.86PDD227 pKa = 3.81TEE229 pKa = 4.3GG230 pKa = 3.67
MM1 pKa = 8.26DD2 pKa = 4.53ISQINSYY9 pKa = 10.88SSASTKK15 pKa = 10.23RR16 pKa = 11.84SSANSSSSLSIEE28 pKa = 4.01QFLSLLAAQLSNQDD42 pKa = 3.67VLNPTQDD49 pKa = 3.52TEE51 pKa = 5.07FVAQMAQFTSLQALEE66 pKa = 4.79NLNQYY71 pKa = 11.06ASYY74 pKa = 10.74QYY76 pKa = 11.0GSSLIGKK83 pKa = 8.42KK84 pKa = 10.48VSVAMYY90 pKa = 10.08DD91 pKa = 3.31STGKK95 pKa = 10.21YY96 pKa = 8.96VSDD99 pKa = 3.51MGVVTQADD107 pKa = 4.33YY108 pKa = 11.61SSGDD112 pKa = 3.2TTVTVNGRR120 pKa = 11.84SYY122 pKa = 11.34GIANIMEE129 pKa = 4.38VFNDD133 pKa = 3.49ASYY136 pKa = 11.6GSYY139 pKa = 10.34QYY141 pKa = 11.32AASLVGKK148 pKa = 9.37KK149 pKa = 8.76VQVTGYY155 pKa = 9.86DD156 pKa = 3.28VNGNVVEE163 pKa = 4.16NTGIVSSCTFASGEE177 pKa = 3.99AMVVVDD183 pKa = 4.02GQKK186 pKa = 8.83YY187 pKa = 6.98TLSAVRR193 pKa = 11.84QVLSQNADD201 pKa = 3.6PEE203 pKa = 4.38PTDD206 pKa = 4.51PEE208 pKa = 4.42TSDD211 pKa = 3.54DD212 pKa = 3.84TTVPDD217 pKa = 4.03TDD219 pKa = 4.0SSGSTDD225 pKa = 3.86PDD227 pKa = 3.81TEE229 pKa = 4.3GG230 pKa = 3.67
Molecular weight: 24.37 kDa
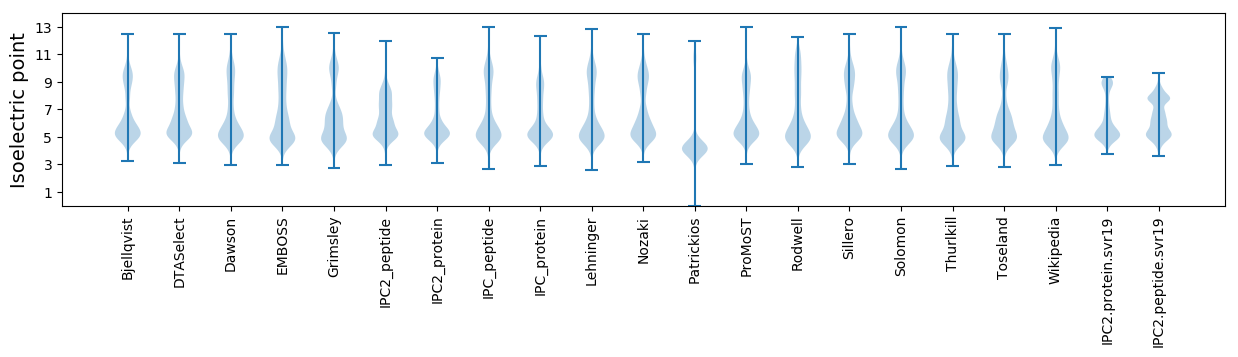
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J0MW05|J0MW05_9CLOT Uncharacterized protein OS=Clostridium sp. MSTE9 OX=1105031 GN=HMPREF1141_2091 PE=4 SV=1
MM1 pKa = 7.31GSSRR5 pKa = 11.84RR6 pKa = 11.84STLAIFACAVSLLLAALLALSLGAVRR32 pKa = 11.84LSLTEE37 pKa = 3.8LFYY40 pKa = 11.22GLKK43 pKa = 10.39EE44 pKa = 3.78IDD46 pKa = 3.83SPAGRR51 pKa = 11.84ILLHH55 pKa = 5.56VRR57 pKa = 11.84VPRR60 pKa = 11.84LLAAALAGAGLSVSGVLIQTVLQNPLASPGIIGVNAGAGLAVALCMALLPGVVLVLPFAAFLGALASVLLVYY132 pKa = 10.16GIARR136 pKa = 11.84RR137 pKa = 11.84AGASRR142 pKa = 11.84ITLVLSGVALSSLMTAGINTLVTVFPDD169 pKa = 2.86IMTGLRR175 pKa = 11.84DD176 pKa = 3.7FQNGGFEE183 pKa = 4.34GVSTGILRR191 pKa = 11.84PAGLVILFGLLVSLLFAGEE210 pKa = 4.34LDD212 pKa = 3.66VLGLGEE218 pKa = 4.23ITASSLGLPVRR229 pKa = 11.84FYY231 pKa = 11.42RR232 pKa = 11.84FLFLALAAALAGAAVSFAGLLGFVGLIVPHH262 pKa = 5.96MARR265 pKa = 11.84FLLKK269 pKa = 10.71GSGKK273 pKa = 10.23RR274 pKa = 11.84SFLLLSALLGAAFLILCDD292 pKa = 3.54TAARR296 pKa = 11.84TAFAPFEE303 pKa = 4.39LPVGILVAYY312 pKa = 10.28LGVPFFLWLLFRR324 pKa = 11.84EE325 pKa = 3.94RR326 pKa = 11.84RR327 pKa = 11.84KK328 pKa = 10.38RR329 pKa = 11.84HH330 pKa = 5.54DD331 pKa = 3.17
MM1 pKa = 7.31GSSRR5 pKa = 11.84RR6 pKa = 11.84STLAIFACAVSLLLAALLALSLGAVRR32 pKa = 11.84LSLTEE37 pKa = 3.8LFYY40 pKa = 11.22GLKK43 pKa = 10.39EE44 pKa = 3.78IDD46 pKa = 3.83SPAGRR51 pKa = 11.84ILLHH55 pKa = 5.56VRR57 pKa = 11.84VPRR60 pKa = 11.84LLAAALAGAGLSVSGVLIQTVLQNPLASPGIIGVNAGAGLAVALCMALLPGVVLVLPFAAFLGALASVLLVYY132 pKa = 10.16GIARR136 pKa = 11.84RR137 pKa = 11.84AGASRR142 pKa = 11.84ITLVLSGVALSSLMTAGINTLVTVFPDD169 pKa = 2.86IMTGLRR175 pKa = 11.84DD176 pKa = 3.7FQNGGFEE183 pKa = 4.34GVSTGILRR191 pKa = 11.84PAGLVILFGLLVSLLFAGEE210 pKa = 4.34LDD212 pKa = 3.66VLGLGEE218 pKa = 4.23ITASSLGLPVRR229 pKa = 11.84FYY231 pKa = 11.42RR232 pKa = 11.84FLFLALAAALAGAAVSFAGLLGFVGLIVPHH262 pKa = 5.96MARR265 pKa = 11.84FLLKK269 pKa = 10.71GSGKK273 pKa = 10.23RR274 pKa = 11.84SFLLLSALLGAAFLILCDD292 pKa = 3.54TAARR296 pKa = 11.84TAFAPFEE303 pKa = 4.39LPVGILVAYY312 pKa = 10.28LGVPFFLWLLFRR324 pKa = 11.84EE325 pKa = 3.94RR326 pKa = 11.84RR327 pKa = 11.84KK328 pKa = 10.38RR329 pKa = 11.84HH330 pKa = 5.54DD331 pKa = 3.17
Molecular weight: 34.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
987318 |
37 |
2204 |
283.9 |
31.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.729 ± 0.05 | 1.482 ± 0.018 |
5.045 ± 0.032 | 6.698 ± 0.045 |
4.136 ± 0.033 | 7.511 ± 0.043 |
1.791 ± 0.019 | 6.674 ± 0.037 |
5.37 ± 0.033 | 9.903 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.9 ± 0.022 | 3.732 ± 0.03 |
4.014 ± 0.03 | 3.845 ± 0.03 |
5.064 ± 0.041 | 6.372 ± 0.041 |
5.356 ± 0.034 | 7.095 ± 0.035 |
0.881 ± 0.013 | 3.403 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
