
Vesicular stomatitis virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Vesiculovirus; unclassified Vesiculovirus
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
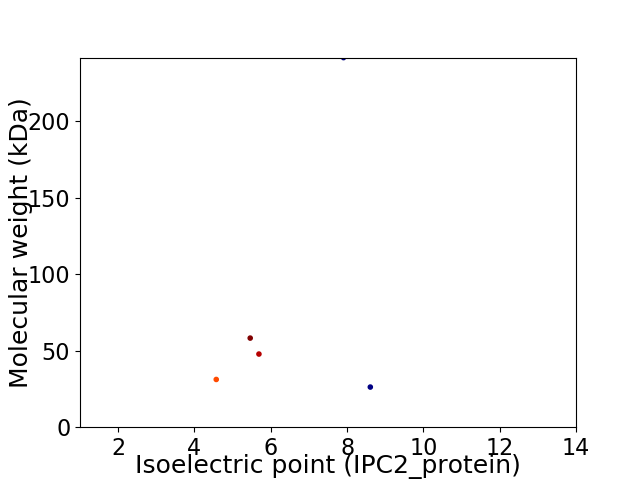
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A110ASW0|A0A110ASW0_9RHAB GDP polyribonucleotidyltransferase OS=Vesicular stomatitis virus OX=11276 PE=3 SV=1
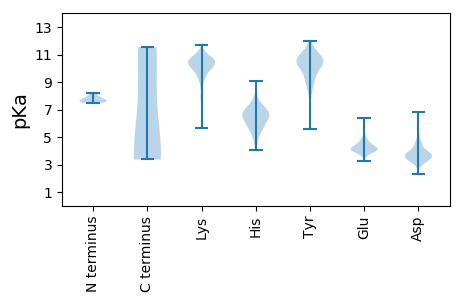
MM1 pKa = 8.21DD2 pKa = 5.84SIDD5 pKa = 3.68RR6 pKa = 11.84LKK8 pKa = 10.32TYY10 pKa = 10.46LATYY14 pKa = 10.48DD15 pKa = 4.05NLDD18 pKa = 4.09SALQDD23 pKa = 3.42ANEE26 pKa = 4.16SEE28 pKa = 4.3EE29 pKa = 4.06RR30 pKa = 11.84RR31 pKa = 11.84EE32 pKa = 4.2DD33 pKa = 4.49KK34 pKa = 10.96YY35 pKa = 11.59LQDD38 pKa = 5.98LFIEE42 pKa = 4.58DD43 pKa = 4.04QGDD46 pKa = 3.64KK47 pKa = 8.78PTPSYY52 pKa = 10.19YY53 pKa = 10.06QEE55 pKa = 4.54EE56 pKa = 4.47EE57 pKa = 4.43SSDD60 pKa = 3.45SDD62 pKa = 3.11TDD64 pKa = 3.66YY65 pKa = 11.37NAEE68 pKa = 4.28HH69 pKa = 6.68LTMLSPDD76 pKa = 3.2EE77 pKa = 6.02RR78 pKa = 11.84IDD80 pKa = 3.24KK81 pKa = 10.3WEE83 pKa = 3.96EE84 pKa = 3.59DD85 pKa = 3.67LPEE88 pKa = 4.77LEE90 pKa = 5.82KK91 pKa = 10.31IDD93 pKa = 5.26DD94 pKa = 5.31DD95 pKa = 4.61IPVTFSDD102 pKa = 3.17WTQPVMKK109 pKa = 10.35GNGGEE114 pKa = 4.13KK115 pKa = 10.02SLSLFPPVGLTRR127 pKa = 11.84VQTDD131 pKa = 2.32QWMKK135 pKa = 10.18TIEE138 pKa = 4.15AVCEE142 pKa = 4.01SSKK145 pKa = 10.81YY146 pKa = 9.41WNLSEE151 pKa = 4.35CRR153 pKa = 11.84IMNSGNCLILKK164 pKa = 9.13GRR166 pKa = 11.84VMTPDD171 pKa = 3.78CSSSTKK177 pKa = 8.59SQKK180 pKa = 10.89SMQSSEE186 pKa = 4.46SLSSSHH192 pKa = 6.88SPGPAPKK199 pKa = 8.9VTEE202 pKa = 4.32STSLWDD208 pKa = 3.81LKK210 pKa = 10.34STEE213 pKa = 3.93VQLISKK219 pKa = 9.9RR220 pKa = 11.84AGVKK224 pKa = 10.79DD225 pKa = 3.63MMVKK229 pKa = 9.46LTDD232 pKa = 4.01FFRR235 pKa = 11.84SEE237 pKa = 3.89DD238 pKa = 3.69EE239 pKa = 4.49YY240 pKa = 11.79YY241 pKa = 10.76SVCPEE246 pKa = 4.24GAPDD250 pKa = 3.98LMGAIIMGLKK260 pKa = 9.59HH261 pKa = 6.63KK262 pKa = 10.68KK263 pKa = 10.02LFNQARR269 pKa = 11.84MKK271 pKa = 10.8YY272 pKa = 9.76RR273 pKa = 11.84LL274 pKa = 3.43
MM1 pKa = 8.21DD2 pKa = 5.84SIDD5 pKa = 3.68RR6 pKa = 11.84LKK8 pKa = 10.32TYY10 pKa = 10.46LATYY14 pKa = 10.48DD15 pKa = 4.05NLDD18 pKa = 4.09SALQDD23 pKa = 3.42ANEE26 pKa = 4.16SEE28 pKa = 4.3EE29 pKa = 4.06RR30 pKa = 11.84RR31 pKa = 11.84EE32 pKa = 4.2DD33 pKa = 4.49KK34 pKa = 10.96YY35 pKa = 11.59LQDD38 pKa = 5.98LFIEE42 pKa = 4.58DD43 pKa = 4.04QGDD46 pKa = 3.64KK47 pKa = 8.78PTPSYY52 pKa = 10.19YY53 pKa = 10.06QEE55 pKa = 4.54EE56 pKa = 4.47EE57 pKa = 4.43SSDD60 pKa = 3.45SDD62 pKa = 3.11TDD64 pKa = 3.66YY65 pKa = 11.37NAEE68 pKa = 4.28HH69 pKa = 6.68LTMLSPDD76 pKa = 3.2EE77 pKa = 6.02RR78 pKa = 11.84IDD80 pKa = 3.24KK81 pKa = 10.3WEE83 pKa = 3.96EE84 pKa = 3.59DD85 pKa = 3.67LPEE88 pKa = 4.77LEE90 pKa = 5.82KK91 pKa = 10.31IDD93 pKa = 5.26DD94 pKa = 5.31DD95 pKa = 4.61IPVTFSDD102 pKa = 3.17WTQPVMKK109 pKa = 10.35GNGGEE114 pKa = 4.13KK115 pKa = 10.02SLSLFPPVGLTRR127 pKa = 11.84VQTDD131 pKa = 2.32QWMKK135 pKa = 10.18TIEE138 pKa = 4.15AVCEE142 pKa = 4.01SSKK145 pKa = 10.81YY146 pKa = 9.41WNLSEE151 pKa = 4.35CRR153 pKa = 11.84IMNSGNCLILKK164 pKa = 9.13GRR166 pKa = 11.84VMTPDD171 pKa = 3.78CSSSTKK177 pKa = 8.59SQKK180 pKa = 10.89SMQSSEE186 pKa = 4.46SLSSSHH192 pKa = 6.88SPGPAPKK199 pKa = 8.9VTEE202 pKa = 4.32STSLWDD208 pKa = 3.81LKK210 pKa = 10.34STEE213 pKa = 3.93VQLISKK219 pKa = 9.9RR220 pKa = 11.84AGVKK224 pKa = 10.79DD225 pKa = 3.63MMVKK229 pKa = 9.46LTDD232 pKa = 4.01FFRR235 pKa = 11.84SEE237 pKa = 3.89DD238 pKa = 3.69EE239 pKa = 4.49YY240 pKa = 11.79YY241 pKa = 10.76SVCPEE246 pKa = 4.24GAPDD250 pKa = 3.98LMGAIIMGLKK260 pKa = 9.59HH261 pKa = 6.63KK262 pKa = 10.68KK263 pKa = 10.02LFNQARR269 pKa = 11.84MKK271 pKa = 10.8YY272 pKa = 9.76RR273 pKa = 11.84LL274 pKa = 3.43
Molecular weight: 31.2 kDa
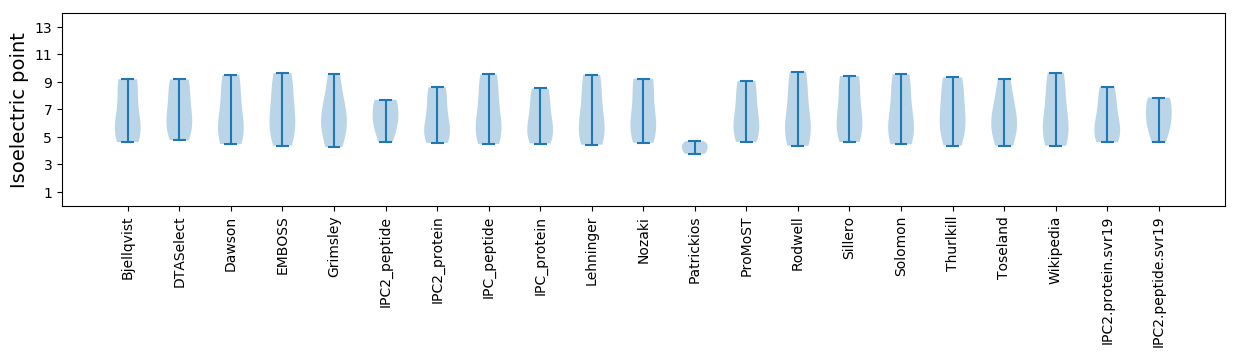
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Y0IXQ8|A0A0Y0IXQ8_9RHAB Nucleocapsid protein OS=Vesicular stomatitis virus OX=11276 PE=3 SV=1
MM1 pKa = 7.72SSFKK5 pKa = 10.64KK6 pKa = 10.4ILGLSSRR13 pKa = 11.84SHH15 pKa = 6.23KK16 pKa = 10.16KK17 pKa = 9.77SKK19 pKa = 10.66KK20 pKa = 9.96LGLPPPYY27 pKa = 10.09DD28 pKa = 3.46EE29 pKa = 4.92SSPMEE34 pKa = 4.19IQPSAPLSNDD44 pKa = 2.95FFGMEE49 pKa = 5.58DD50 pKa = 3.18MDD52 pKa = 6.38LYY54 pKa = 11.49DD55 pKa = 5.09KK56 pKa = 11.34DD57 pKa = 3.49SLRR60 pKa = 11.84YY61 pKa = 8.84EE62 pKa = 4.36KK63 pKa = 10.58FRR65 pKa = 11.84FMLKK69 pKa = 7.95MTVRR73 pKa = 11.84SNKK76 pKa = 9.31PFRR79 pKa = 11.84SYY81 pKa = 11.86DD82 pKa = 3.46DD83 pKa = 3.16VTAAVSQWDD92 pKa = 3.41NSYY95 pKa = 10.93IGMVGKK101 pKa = 10.57RR102 pKa = 11.84PFYY105 pKa = 10.59KK106 pKa = 9.95IIALIGSSHH115 pKa = 6.42LQATPAVLADD125 pKa = 4.27LNQPEE130 pKa = 4.8YY131 pKa = 10.9YY132 pKa = 9.36ATLTGRR138 pKa = 11.84CFLPHH143 pKa = 7.26RR144 pKa = 11.84LGLIPPMFNVSEE156 pKa = 4.38TFRR159 pKa = 11.84KK160 pKa = 9.19PFNIGIYY167 pKa = 10.12KK168 pKa = 8.76GTLDD172 pKa = 3.54FTFTVSDD179 pKa = 3.91DD180 pKa = 3.65EE181 pKa = 5.1SNEE184 pKa = 4.02KK185 pKa = 10.14VPHH188 pKa = 5.27IWEE191 pKa = 4.17YY192 pKa = 11.07MNPKK196 pKa = 9.39YY197 pKa = 10.39QSQIQKK203 pKa = 10.19EE204 pKa = 4.28GLKK207 pKa = 10.63FGLILSKK214 pKa = 10.55KK215 pKa = 9.05ATGTWVLDD223 pKa = 3.31QLSPFKK229 pKa = 11.04
MM1 pKa = 7.72SSFKK5 pKa = 10.64KK6 pKa = 10.4ILGLSSRR13 pKa = 11.84SHH15 pKa = 6.23KK16 pKa = 10.16KK17 pKa = 9.77SKK19 pKa = 10.66KK20 pKa = 9.96LGLPPPYY27 pKa = 10.09DD28 pKa = 3.46EE29 pKa = 4.92SSPMEE34 pKa = 4.19IQPSAPLSNDD44 pKa = 2.95FFGMEE49 pKa = 5.58DD50 pKa = 3.18MDD52 pKa = 6.38LYY54 pKa = 11.49DD55 pKa = 5.09KK56 pKa = 11.34DD57 pKa = 3.49SLRR60 pKa = 11.84YY61 pKa = 8.84EE62 pKa = 4.36KK63 pKa = 10.58FRR65 pKa = 11.84FMLKK69 pKa = 7.95MTVRR73 pKa = 11.84SNKK76 pKa = 9.31PFRR79 pKa = 11.84SYY81 pKa = 11.86DD82 pKa = 3.46DD83 pKa = 3.16VTAAVSQWDD92 pKa = 3.41NSYY95 pKa = 10.93IGMVGKK101 pKa = 10.57RR102 pKa = 11.84PFYY105 pKa = 10.59KK106 pKa = 9.95IIALIGSSHH115 pKa = 6.42LQATPAVLADD125 pKa = 4.27LNQPEE130 pKa = 4.8YY131 pKa = 10.9YY132 pKa = 9.36ATLTGRR138 pKa = 11.84CFLPHH143 pKa = 7.26RR144 pKa = 11.84LGLIPPMFNVSEE156 pKa = 4.38TFRR159 pKa = 11.84KK160 pKa = 9.19PFNIGIYY167 pKa = 10.12KK168 pKa = 8.76GTLDD172 pKa = 3.54FTFTVSDD179 pKa = 3.91DD180 pKa = 3.65EE181 pKa = 5.1SNEE184 pKa = 4.02KK185 pKa = 10.14VPHH188 pKa = 5.27IWEE191 pKa = 4.17YY192 pKa = 11.07MNPKK196 pKa = 9.39YY197 pKa = 10.39QSQIQKK203 pKa = 10.19EE204 pKa = 4.28GLKK207 pKa = 10.63FGLILSKK214 pKa = 10.55KK215 pKa = 9.05ATGTWVLDD223 pKa = 3.31QLSPFKK229 pKa = 11.04
Molecular weight: 26.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3551 |
229 |
2109 |
710.2 |
81.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.168 ± 0.543 | 1.802 ± 0.223 |
6.28 ± 0.7 | 5.829 ± 0.458 |
4.196 ± 0.315 | 6.139 ± 0.461 |
2.534 ± 0.375 | 7.04 ± 0.619 |
7.181 ± 0.378 | 9.434 ± 0.749 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.985 ± 0.289 | 4.14 ± 0.513 |
5.182 ± 0.389 | 3.323 ± 0.252 |
4.9 ± 0.504 | 8.448 ± 0.533 |
5.632 ± 0.46 | 5.041 ± 0.542 |
1.971 ± 0.102 | 3.774 ± 0.265 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
