
Avon-Heathcote Estuary associated circular virus 8
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.61
Get precalculated fractions of proteins
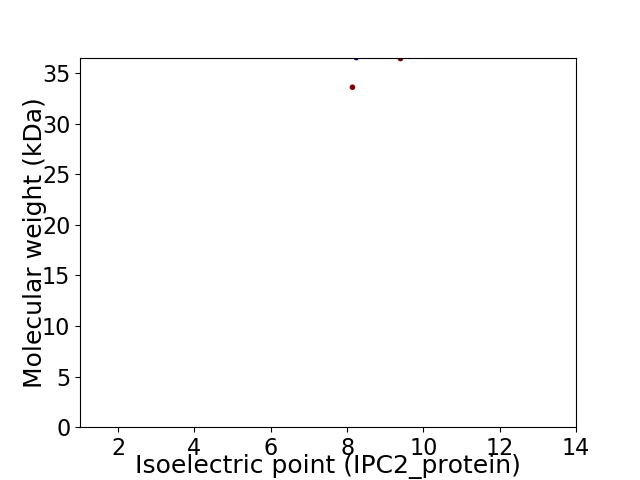
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IAY1|A0A0C5IAY1_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 8 OX=1618259 PE=4 SV=1
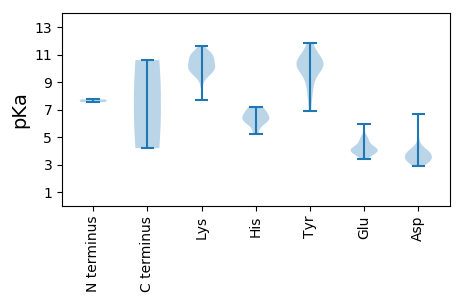
MM1 pKa = 7.54YY2 pKa = 9.93MGDD5 pKa = 3.48SQLAVWDD12 pKa = 4.3FRR14 pKa = 11.84YY15 pKa = 10.11NASTFKK21 pKa = 11.0SEE23 pKa = 4.17SDD25 pKa = 3.19LKK27 pKa = 11.29AVLKK31 pKa = 10.84GIAKK35 pKa = 9.41KK36 pKa = 9.51WVFQLEE42 pKa = 4.37RR43 pKa = 11.84GDD45 pKa = 3.68EE46 pKa = 4.55GYY48 pKa = 10.51EE49 pKa = 4.0HH50 pKa = 6.2YY51 pKa = 10.05QGRR54 pKa = 11.84MSLIKK59 pKa = 10.28GRR61 pKa = 11.84RR62 pKa = 11.84KK63 pKa = 10.2GEE65 pKa = 3.59KK66 pKa = 9.89HH67 pKa = 6.11ILLKK71 pKa = 10.68LFKK74 pKa = 8.77EE75 pKa = 4.57TPPNYY80 pKa = 10.33LEE82 pKa = 4.38PTSKK86 pKa = 9.73TEE88 pKa = 3.9ATKK91 pKa = 11.13GDD93 pKa = 3.7ALYY96 pKa = 10.69VIKK99 pKa = 10.48EE100 pKa = 4.09DD101 pKa = 3.94TRR103 pKa = 11.84LKK105 pKa = 11.0GPWTDD110 pKa = 3.06KK111 pKa = 11.59DD112 pKa = 3.76EE113 pKa = 4.8VKK115 pKa = 10.97VEE117 pKa = 4.06TTQLKK122 pKa = 9.9IFKK125 pKa = 9.9SLEE128 pKa = 3.41LRR130 pKa = 11.84PYY132 pKa = 9.29QRR134 pKa = 11.84KK135 pKa = 9.49LEE137 pKa = 4.11EE138 pKa = 3.82FARR141 pKa = 11.84TFDD144 pKa = 3.35MRR146 pKa = 11.84KK147 pKa = 9.1IDD149 pKa = 4.86IIYY152 pKa = 10.37DD153 pKa = 4.05PIGDD157 pKa = 3.9LGKK160 pKa = 10.69SLFSEE165 pKa = 3.62WMEE168 pKa = 3.94YY169 pKa = 10.63EE170 pKa = 4.18GLAEE174 pKa = 4.95EE175 pKa = 4.3IPPYY179 pKa = 10.86RR180 pKa = 11.84LMDD183 pKa = 6.69DD184 pKa = 3.62IFQWVATRR192 pKa = 11.84PIKK195 pKa = 10.07PCYY198 pKa = 8.94IVDD201 pKa = 3.69MPRR204 pKa = 11.84GMKK207 pKa = 9.53KK208 pKa = 10.41DD209 pKa = 3.5KK210 pKa = 11.36LGDD213 pKa = 3.75FYY215 pKa = 11.81SGLEE219 pKa = 4.05VIKK222 pKa = 11.18NGVAYY227 pKa = 10.04DD228 pKa = 3.46KK229 pKa = 10.61RR230 pKa = 11.84YY231 pKa = 9.74NATKK235 pKa = 10.15IRR237 pKa = 11.84FNRR240 pKa = 11.84PRR242 pKa = 11.84VFVFTNTLPEE252 pKa = 4.13FSLMSKK258 pKa = 10.25DD259 pKa = 3.21RR260 pKa = 11.84WNVWEE265 pKa = 4.04VQEE268 pKa = 5.96DD269 pKa = 3.99YY270 pKa = 11.25DD271 pKa = 3.83ILLRR275 pKa = 11.84NEE277 pKa = 3.96MGTLSMKK284 pKa = 10.6
MM1 pKa = 7.54YY2 pKa = 9.93MGDD5 pKa = 3.48SQLAVWDD12 pKa = 4.3FRR14 pKa = 11.84YY15 pKa = 10.11NASTFKK21 pKa = 11.0SEE23 pKa = 4.17SDD25 pKa = 3.19LKK27 pKa = 11.29AVLKK31 pKa = 10.84GIAKK35 pKa = 9.41KK36 pKa = 9.51WVFQLEE42 pKa = 4.37RR43 pKa = 11.84GDD45 pKa = 3.68EE46 pKa = 4.55GYY48 pKa = 10.51EE49 pKa = 4.0HH50 pKa = 6.2YY51 pKa = 10.05QGRR54 pKa = 11.84MSLIKK59 pKa = 10.28GRR61 pKa = 11.84RR62 pKa = 11.84KK63 pKa = 10.2GEE65 pKa = 3.59KK66 pKa = 9.89HH67 pKa = 6.11ILLKK71 pKa = 10.68LFKK74 pKa = 8.77EE75 pKa = 4.57TPPNYY80 pKa = 10.33LEE82 pKa = 4.38PTSKK86 pKa = 9.73TEE88 pKa = 3.9ATKK91 pKa = 11.13GDD93 pKa = 3.7ALYY96 pKa = 10.69VIKK99 pKa = 10.48EE100 pKa = 4.09DD101 pKa = 3.94TRR103 pKa = 11.84LKK105 pKa = 11.0GPWTDD110 pKa = 3.06KK111 pKa = 11.59DD112 pKa = 3.76EE113 pKa = 4.8VKK115 pKa = 10.97VEE117 pKa = 4.06TTQLKK122 pKa = 9.9IFKK125 pKa = 9.9SLEE128 pKa = 3.41LRR130 pKa = 11.84PYY132 pKa = 9.29QRR134 pKa = 11.84KK135 pKa = 9.49LEE137 pKa = 4.11EE138 pKa = 3.82FARR141 pKa = 11.84TFDD144 pKa = 3.35MRR146 pKa = 11.84KK147 pKa = 9.1IDD149 pKa = 4.86IIYY152 pKa = 10.37DD153 pKa = 4.05PIGDD157 pKa = 3.9LGKK160 pKa = 10.69SLFSEE165 pKa = 3.62WMEE168 pKa = 3.94YY169 pKa = 10.63EE170 pKa = 4.18GLAEE174 pKa = 4.95EE175 pKa = 4.3IPPYY179 pKa = 10.86RR180 pKa = 11.84LMDD183 pKa = 6.69DD184 pKa = 3.62IFQWVATRR192 pKa = 11.84PIKK195 pKa = 10.07PCYY198 pKa = 8.94IVDD201 pKa = 3.69MPRR204 pKa = 11.84GMKK207 pKa = 9.53KK208 pKa = 10.41DD209 pKa = 3.5KK210 pKa = 11.36LGDD213 pKa = 3.75FYY215 pKa = 11.81SGLEE219 pKa = 4.05VIKK222 pKa = 11.18NGVAYY227 pKa = 10.04DD228 pKa = 3.46KK229 pKa = 10.61RR230 pKa = 11.84YY231 pKa = 9.74NATKK235 pKa = 10.15IRR237 pKa = 11.84FNRR240 pKa = 11.84PRR242 pKa = 11.84VFVFTNTLPEE252 pKa = 4.13FSLMSKK258 pKa = 10.25DD259 pKa = 3.21RR260 pKa = 11.84WNVWEE265 pKa = 4.04VQEE268 pKa = 5.96DD269 pKa = 3.99YY270 pKa = 11.25DD271 pKa = 3.83ILLRR275 pKa = 11.84NEE277 pKa = 3.96MGTLSMKK284 pKa = 10.6
Molecular weight: 33.59 kDa
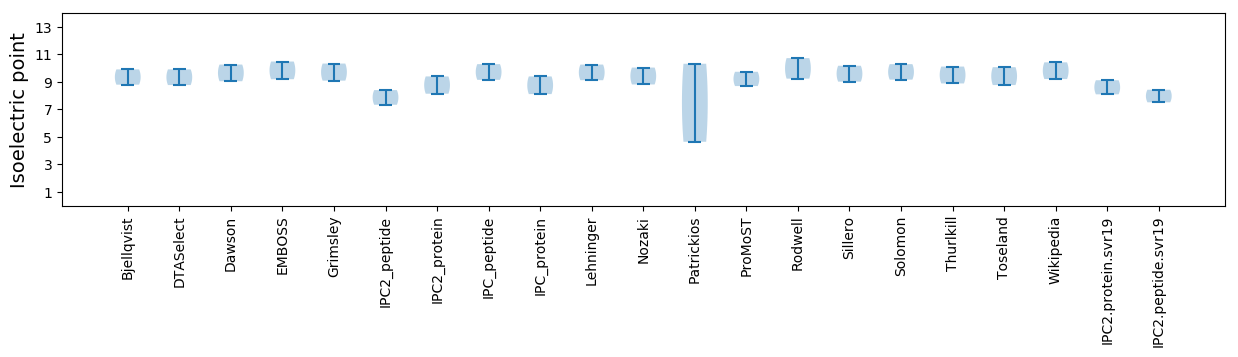
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IAY1|A0A0C5IAY1_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 8 OX=1618259 PE=4 SV=1
MM1 pKa = 7.74PKK3 pKa = 10.29YY4 pKa = 10.15SGPRR8 pKa = 11.84MPSGRR13 pKa = 11.84YY14 pKa = 6.88YY15 pKa = 11.02SKK17 pKa = 10.95RR18 pKa = 11.84KK19 pKa = 9.48AGRR22 pKa = 11.84PKK24 pKa = 10.58GSKK27 pKa = 9.76NKK29 pKa = 9.71PKK31 pKa = 10.67AKK33 pKa = 10.07RR34 pKa = 11.84GGLNKK39 pKa = 10.13TEE41 pKa = 4.24VKK43 pKa = 7.69QTRR46 pKa = 11.84AIAKK50 pKa = 9.71RR51 pKa = 11.84VINSNAEE58 pKa = 3.81SKK60 pKa = 10.94YY61 pKa = 10.32FDD63 pKa = 3.93VRR65 pKa = 11.84TIDD68 pKa = 3.41QLSGSGGVGIVPTPARR84 pKa = 11.84LAYY87 pKa = 7.85TQLYY91 pKa = 8.2VQGFAVGEE99 pKa = 4.07NTIGGTALTYY109 pKa = 9.62GQTVIEE115 pKa = 5.03PINHH119 pKa = 5.22GRR121 pKa = 11.84IHH123 pKa = 7.19PSGNSDD129 pKa = 3.29NQNLEE134 pKa = 3.67GQYY137 pKa = 11.59AMPSLSITTFDD148 pKa = 3.18IQRR151 pKa = 11.84VIQTSDD157 pKa = 3.08LTLDD161 pKa = 4.26LARR164 pKa = 11.84NQTPFMVRR172 pKa = 11.84VLRR175 pKa = 11.84LVPRR179 pKa = 11.84PQKK182 pKa = 10.79GSSQGLDD189 pKa = 3.33PANDD193 pKa = 3.39AFVNEE198 pKa = 4.74SNQEE202 pKa = 3.84TGVADD207 pKa = 3.43SGFEE211 pKa = 4.02HH212 pKa = 6.53YY213 pKa = 10.45QLLFLKK219 pKa = 10.48ANSKK223 pKa = 10.17KK224 pKa = 10.55YY225 pKa = 8.68QVKK228 pKa = 9.8QDD230 pKa = 3.21FRR232 pKa = 11.84MVLNPPLSTTEE243 pKa = 3.65VAMQGNAGSTAYY255 pKa = 10.3SVANISNNFHH265 pKa = 6.24RR266 pKa = 11.84QMTFKK271 pKa = 10.84HH272 pKa = 6.83DD273 pKa = 3.12IGKK276 pKa = 9.92KK277 pKa = 9.98LFYY280 pKa = 10.42DD281 pKa = 3.87NPQSRR286 pKa = 11.84SHH288 pKa = 6.31PTDD291 pKa = 2.87GFKK294 pKa = 11.18NEE296 pKa = 4.97FILFHH301 pKa = 6.96IIPLGTANDD310 pKa = 4.06GGILPTSVRR319 pKa = 11.84IAAKK323 pKa = 10.13AVSTFKK329 pKa = 11.11DD330 pKa = 3.8FF331 pKa = 4.19
MM1 pKa = 7.74PKK3 pKa = 10.29YY4 pKa = 10.15SGPRR8 pKa = 11.84MPSGRR13 pKa = 11.84YY14 pKa = 6.88YY15 pKa = 11.02SKK17 pKa = 10.95RR18 pKa = 11.84KK19 pKa = 9.48AGRR22 pKa = 11.84PKK24 pKa = 10.58GSKK27 pKa = 9.76NKK29 pKa = 9.71PKK31 pKa = 10.67AKK33 pKa = 10.07RR34 pKa = 11.84GGLNKK39 pKa = 10.13TEE41 pKa = 4.24VKK43 pKa = 7.69QTRR46 pKa = 11.84AIAKK50 pKa = 9.71RR51 pKa = 11.84VINSNAEE58 pKa = 3.81SKK60 pKa = 10.94YY61 pKa = 10.32FDD63 pKa = 3.93VRR65 pKa = 11.84TIDD68 pKa = 3.41QLSGSGGVGIVPTPARR84 pKa = 11.84LAYY87 pKa = 7.85TQLYY91 pKa = 8.2VQGFAVGEE99 pKa = 4.07NTIGGTALTYY109 pKa = 9.62GQTVIEE115 pKa = 5.03PINHH119 pKa = 5.22GRR121 pKa = 11.84IHH123 pKa = 7.19PSGNSDD129 pKa = 3.29NQNLEE134 pKa = 3.67GQYY137 pKa = 11.59AMPSLSITTFDD148 pKa = 3.18IQRR151 pKa = 11.84VIQTSDD157 pKa = 3.08LTLDD161 pKa = 4.26LARR164 pKa = 11.84NQTPFMVRR172 pKa = 11.84VLRR175 pKa = 11.84LVPRR179 pKa = 11.84PQKK182 pKa = 10.79GSSQGLDD189 pKa = 3.33PANDD193 pKa = 3.39AFVNEE198 pKa = 4.74SNQEE202 pKa = 3.84TGVADD207 pKa = 3.43SGFEE211 pKa = 4.02HH212 pKa = 6.53YY213 pKa = 10.45QLLFLKK219 pKa = 10.48ANSKK223 pKa = 10.17KK224 pKa = 10.55YY225 pKa = 8.68QVKK228 pKa = 9.8QDD230 pKa = 3.21FRR232 pKa = 11.84MVLNPPLSTTEE243 pKa = 3.65VAMQGNAGSTAYY255 pKa = 10.3SVANISNNFHH265 pKa = 6.24RR266 pKa = 11.84QMTFKK271 pKa = 10.84HH272 pKa = 6.83DD273 pKa = 3.12IGKK276 pKa = 9.92KK277 pKa = 9.98LFYY280 pKa = 10.42DD281 pKa = 3.87NPQSRR286 pKa = 11.84SHH288 pKa = 6.31PTDD291 pKa = 2.87GFKK294 pKa = 11.18NEE296 pKa = 4.97FILFHH301 pKa = 6.96IIPLGTANDD310 pKa = 4.06GGILPTSVRR319 pKa = 11.84IAAKK323 pKa = 10.13AVSTFKK329 pKa = 11.11DD330 pKa = 3.8FF331 pKa = 4.19
Molecular weight: 36.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
615 |
284 |
331 |
307.5 |
35.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.528 ± 1.045 | 0.163 ± 0.12 |
5.854 ± 0.972 | 5.528 ± 1.844 |
4.878 ± 0.033 | 7.48 ± 0.943 |
1.463 ± 0.479 | 5.528 ± 0.066 |
8.618 ± 1.228 | 7.805 ± 0.852 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.927 ± 0.597 | 4.878 ± 1.301 |
5.366 ± 0.498 | 4.228 ± 1.112 |
6.179 ± 0.323 | 6.341 ± 1.113 |
6.179 ± 0.566 | 5.528 ± 0.378 |
1.138 ± 0.837 | 4.39 ± 0.563 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
