
Leporid alphaherpesvirus 4
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Simplexvirus
Average proteome isoelectric point is 7.43
Get precalculated fractions of proteins
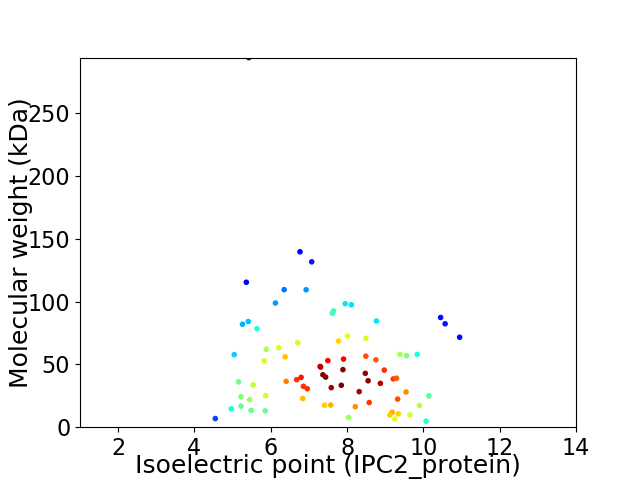
Virtual 2D-PAGE plot for 79 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J9R053|J9R053_9ALPH DNA packaging tegument protein UL17 OS=Leporid alphaherpesvirus 4 OX=481315 GN=UL17a PE=4 SV=1
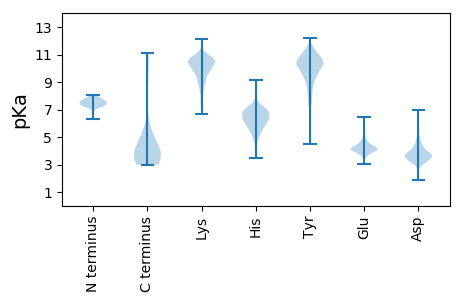
MM1 pKa = 7.51VSLTSDD7 pKa = 3.25EE8 pKa = 4.3FDD10 pKa = 3.56QVDD13 pKa = 3.92ISPTSSDD20 pKa = 2.87EE21 pKa = 4.5FYY23 pKa = 10.98LVPAEE28 pKa = 4.04MQKK31 pKa = 10.99KK32 pKa = 8.77YY33 pKa = 6.99MTGRR37 pKa = 11.84GYY39 pKa = 9.14TRR41 pKa = 11.84IEE43 pKa = 4.08SSPEE47 pKa = 3.63KK48 pKa = 10.23PADD51 pKa = 3.98RR52 pKa = 11.84PSSSPVSPQPP62 pKa = 3.37
MM1 pKa = 7.51VSLTSDD7 pKa = 3.25EE8 pKa = 4.3FDD10 pKa = 3.56QVDD13 pKa = 3.92ISPTSSDD20 pKa = 2.87EE21 pKa = 4.5FYY23 pKa = 10.98LVPAEE28 pKa = 4.04MQKK31 pKa = 10.99KK32 pKa = 8.77YY33 pKa = 6.99MTGRR37 pKa = 11.84GYY39 pKa = 9.14TRR41 pKa = 11.84IEE43 pKa = 4.08SSPEE47 pKa = 3.63KK48 pKa = 10.23PADD51 pKa = 3.98RR52 pKa = 11.84PSSSPVSPQPP62 pKa = 3.37
Molecular weight: 6.9 kDa
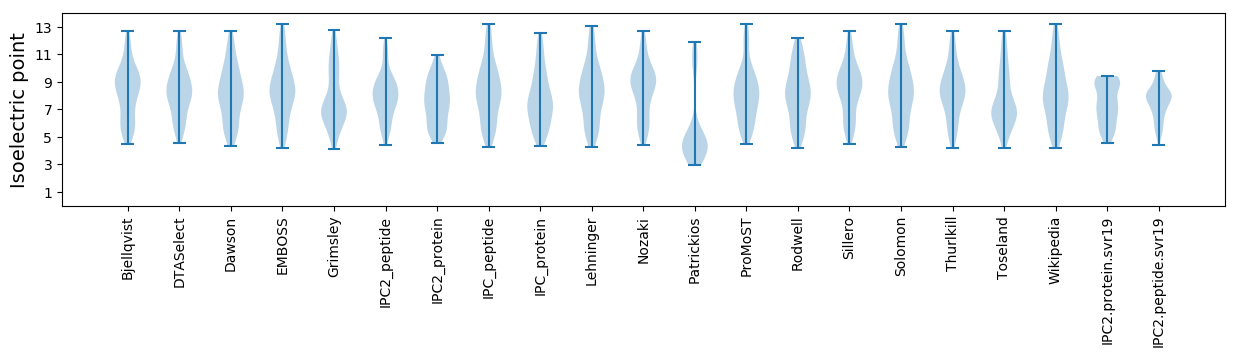
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J9QYL4|J9QYL4_9ALPH Uracil-DNA glycosylase OS=Leporid alphaherpesvirus 4 OX=481315 GN=UL2 PE=3 SV=1
MM1 pKa = 7.5PAHH4 pKa = 6.95RR5 pKa = 11.84PCAAVRR11 pKa = 11.84EE12 pKa = 4.46AAPVTPNPVLRR23 pKa = 11.84GTRR26 pKa = 11.84HH27 pKa = 4.67SRR29 pKa = 11.84VYY31 pKa = 10.33RR32 pKa = 11.84VGPAEE37 pKa = 3.96GVPFARR43 pKa = 11.84HH44 pKa = 4.97TSASHH49 pKa = 6.78RR50 pKa = 11.84PPAPLPARR58 pKa = 11.84LTAGLFDD65 pKa = 4.38TLNLTAPRR73 pKa = 11.84VGDD76 pKa = 3.69PGRR79 pKa = 11.84VQVIVAEE86 pKa = 4.33RR87 pKa = 11.84DD88 pKa = 3.1RR89 pKa = 11.84LTRR92 pKa = 11.84DD93 pKa = 3.11VPSNDD98 pKa = 2.43EE99 pKa = 4.27RR100 pKa = 11.84IARR103 pKa = 11.84RR104 pKa = 11.84CVRR107 pKa = 11.84GPPHH111 pKa = 6.95PGGPGTAAISCASPRR126 pKa = 11.84RR127 pKa = 11.84AGGRR131 pKa = 11.84VPIATGPPSGGGVGSLRR148 pKa = 11.84RR149 pKa = 11.84GGGSLGTSRR158 pKa = 11.84RR159 pKa = 11.84HH160 pKa = 4.7SRR162 pKa = 11.84LVRR165 pKa = 11.84GVASAAPGASSAPRR179 pKa = 11.84GPPPHH184 pKa = 6.88SPWAPPPHH192 pKa = 6.64RR193 pKa = 11.84RR194 pKa = 11.84LPPVRR199 pKa = 11.84PLPSRR204 pKa = 11.84GARR207 pKa = 11.84RR208 pKa = 11.84ARR210 pKa = 11.84KK211 pKa = 8.44PPPAARR217 pKa = 11.84RR218 pKa = 11.84PARR221 pKa = 11.84RR222 pKa = 11.84GRR224 pKa = 11.84PGGGARR230 pKa = 11.84ARR232 pKa = 11.84IPPGARR238 pKa = 11.84PDD240 pKa = 3.74PAPPPRR246 pKa = 11.84APPRR250 pKa = 11.84VSRR253 pKa = 11.84GPPPGARR260 pKa = 11.84GVAQSSRR267 pKa = 11.84GRR269 pKa = 11.84GRR271 pKa = 11.84GAQTEE276 pKa = 4.43EE277 pKa = 4.02SRR279 pKa = 11.84SPCSCEE285 pKa = 3.6GQAALLGPSAPPRR298 pKa = 11.84KK299 pKa = 9.81GPPPRR304 pKa = 11.84GARR307 pKa = 11.84APRR310 pKa = 11.84PSAAPPPTGPWRR322 pKa = 11.84GGPRR326 pKa = 11.84RR327 pKa = 11.84TPSGCSPAAAGPRR340 pKa = 11.84PPPARR345 pKa = 11.84AARR348 pKa = 11.84PRR350 pKa = 11.84GPARR354 pKa = 11.84GARR357 pKa = 11.84PRR359 pKa = 11.84RR360 pKa = 11.84TRR362 pKa = 11.84AGPRR366 pKa = 11.84ARR368 pKa = 11.84PRR370 pKa = 11.84AISARR375 pKa = 11.84RR376 pKa = 11.84RR377 pKa = 11.84AAGRR381 pKa = 11.84GTRR384 pKa = 11.84GGARR388 pKa = 11.84RR389 pKa = 11.84RR390 pKa = 11.84APSARR395 pKa = 11.84RR396 pKa = 11.84RR397 pKa = 11.84GASAPRR403 pKa = 11.84RR404 pKa = 11.84PGSPRR409 pKa = 11.84RR410 pKa = 11.84PGPWPPRR417 pKa = 11.84RR418 pKa = 11.84RR419 pKa = 11.84WWRR422 pKa = 11.84RR423 pKa = 11.84RR424 pKa = 11.84ASRR427 pKa = 11.84ARR429 pKa = 11.84ARR431 pKa = 11.84RR432 pKa = 11.84RR433 pKa = 11.84DD434 pKa = 3.36GTRR437 pKa = 11.84AARR440 pKa = 11.84GARR443 pKa = 11.84AWPRR447 pKa = 11.84RR448 pKa = 11.84RR449 pKa = 11.84ACARR453 pKa = 11.84RR454 pKa = 11.84SARR457 pKa = 11.84RR458 pKa = 11.84RR459 pKa = 11.84RR460 pKa = 11.84GRR462 pKa = 11.84GAGAARR468 pKa = 11.84RR469 pKa = 11.84GSRR472 pKa = 11.84ARR474 pKa = 11.84ARR476 pKa = 11.84PRR478 pKa = 11.84RR479 pKa = 11.84ARR481 pKa = 11.84RR482 pKa = 11.84ARR484 pKa = 11.84ARR486 pKa = 11.84RR487 pKa = 11.84RR488 pKa = 11.84RR489 pKa = 11.84APRR492 pKa = 11.84ARR494 pKa = 11.84RR495 pKa = 11.84RR496 pKa = 11.84AGRR499 pKa = 11.84RR500 pKa = 11.84RR501 pKa = 11.84RR502 pKa = 11.84ASARR506 pKa = 11.84RR507 pKa = 11.84TGPRR511 pKa = 11.84GAPRR515 pKa = 11.84RR516 pKa = 11.84PGTRR520 pKa = 11.84AARR523 pKa = 11.84RR524 pKa = 11.84RR525 pKa = 11.84PGRR528 pKa = 11.84PRR530 pKa = 11.84AXASAGTALTTTSFRR545 pKa = 11.84PAAPASSPRR554 pKa = 11.84NSTAPAKK561 pKa = 10.33ARR563 pKa = 11.84SRR565 pKa = 11.84VEE567 pKa = 3.83SSSTPRR573 pKa = 11.84AFSAEE578 pKa = 4.09TSGGPVQLPAQAAEE592 pKa = 4.23SGVQRR597 pKa = 11.84RR598 pKa = 11.84LASAASRR605 pKa = 11.84NEE607 pKa = 3.85SPPRR611 pKa = 11.84SVVXLXGARR620 pKa = 11.84ARR622 pKa = 11.84ARR624 pKa = 11.84RR625 pKa = 11.84APRR628 pKa = 11.84RR629 pKa = 11.84AAARR633 pKa = 11.84RR634 pKa = 11.84ARR636 pKa = 11.84ARR638 pKa = 11.84ARR640 pKa = 11.84RR641 pKa = 11.84PGLAPGASTRR651 pKa = 11.84RR652 pKa = 11.84RR653 pKa = 11.84GAAGPRR659 pKa = 11.84RR660 pKa = 11.84AAGRR664 pKa = 11.84KK665 pKa = 9.13AGAAPALLACSRR677 pKa = 11.84ASSGAA682 pKa = 3.27
MM1 pKa = 7.5PAHH4 pKa = 6.95RR5 pKa = 11.84PCAAVRR11 pKa = 11.84EE12 pKa = 4.46AAPVTPNPVLRR23 pKa = 11.84GTRR26 pKa = 11.84HH27 pKa = 4.67SRR29 pKa = 11.84VYY31 pKa = 10.33RR32 pKa = 11.84VGPAEE37 pKa = 3.96GVPFARR43 pKa = 11.84HH44 pKa = 4.97TSASHH49 pKa = 6.78RR50 pKa = 11.84PPAPLPARR58 pKa = 11.84LTAGLFDD65 pKa = 4.38TLNLTAPRR73 pKa = 11.84VGDD76 pKa = 3.69PGRR79 pKa = 11.84VQVIVAEE86 pKa = 4.33RR87 pKa = 11.84DD88 pKa = 3.1RR89 pKa = 11.84LTRR92 pKa = 11.84DD93 pKa = 3.11VPSNDD98 pKa = 2.43EE99 pKa = 4.27RR100 pKa = 11.84IARR103 pKa = 11.84RR104 pKa = 11.84CVRR107 pKa = 11.84GPPHH111 pKa = 6.95PGGPGTAAISCASPRR126 pKa = 11.84RR127 pKa = 11.84AGGRR131 pKa = 11.84VPIATGPPSGGGVGSLRR148 pKa = 11.84RR149 pKa = 11.84GGGSLGTSRR158 pKa = 11.84RR159 pKa = 11.84HH160 pKa = 4.7SRR162 pKa = 11.84LVRR165 pKa = 11.84GVASAAPGASSAPRR179 pKa = 11.84GPPPHH184 pKa = 6.88SPWAPPPHH192 pKa = 6.64RR193 pKa = 11.84RR194 pKa = 11.84LPPVRR199 pKa = 11.84PLPSRR204 pKa = 11.84GARR207 pKa = 11.84RR208 pKa = 11.84ARR210 pKa = 11.84KK211 pKa = 8.44PPPAARR217 pKa = 11.84RR218 pKa = 11.84PARR221 pKa = 11.84RR222 pKa = 11.84GRR224 pKa = 11.84PGGGARR230 pKa = 11.84ARR232 pKa = 11.84IPPGARR238 pKa = 11.84PDD240 pKa = 3.74PAPPPRR246 pKa = 11.84APPRR250 pKa = 11.84VSRR253 pKa = 11.84GPPPGARR260 pKa = 11.84GVAQSSRR267 pKa = 11.84GRR269 pKa = 11.84GRR271 pKa = 11.84GAQTEE276 pKa = 4.43EE277 pKa = 4.02SRR279 pKa = 11.84SPCSCEE285 pKa = 3.6GQAALLGPSAPPRR298 pKa = 11.84KK299 pKa = 9.81GPPPRR304 pKa = 11.84GARR307 pKa = 11.84APRR310 pKa = 11.84PSAAPPPTGPWRR322 pKa = 11.84GGPRR326 pKa = 11.84RR327 pKa = 11.84TPSGCSPAAAGPRR340 pKa = 11.84PPPARR345 pKa = 11.84AARR348 pKa = 11.84PRR350 pKa = 11.84GPARR354 pKa = 11.84GARR357 pKa = 11.84PRR359 pKa = 11.84RR360 pKa = 11.84TRR362 pKa = 11.84AGPRR366 pKa = 11.84ARR368 pKa = 11.84PRR370 pKa = 11.84AISARR375 pKa = 11.84RR376 pKa = 11.84RR377 pKa = 11.84AAGRR381 pKa = 11.84GTRR384 pKa = 11.84GGARR388 pKa = 11.84RR389 pKa = 11.84RR390 pKa = 11.84APSARR395 pKa = 11.84RR396 pKa = 11.84RR397 pKa = 11.84GASAPRR403 pKa = 11.84RR404 pKa = 11.84PGSPRR409 pKa = 11.84RR410 pKa = 11.84PGPWPPRR417 pKa = 11.84RR418 pKa = 11.84RR419 pKa = 11.84WWRR422 pKa = 11.84RR423 pKa = 11.84RR424 pKa = 11.84ASRR427 pKa = 11.84ARR429 pKa = 11.84ARR431 pKa = 11.84RR432 pKa = 11.84RR433 pKa = 11.84DD434 pKa = 3.36GTRR437 pKa = 11.84AARR440 pKa = 11.84GARR443 pKa = 11.84AWPRR447 pKa = 11.84RR448 pKa = 11.84RR449 pKa = 11.84ACARR453 pKa = 11.84RR454 pKa = 11.84SARR457 pKa = 11.84RR458 pKa = 11.84RR459 pKa = 11.84RR460 pKa = 11.84GRR462 pKa = 11.84GAGAARR468 pKa = 11.84RR469 pKa = 11.84GSRR472 pKa = 11.84ARR474 pKa = 11.84ARR476 pKa = 11.84PRR478 pKa = 11.84RR479 pKa = 11.84ARR481 pKa = 11.84RR482 pKa = 11.84ARR484 pKa = 11.84ARR486 pKa = 11.84RR487 pKa = 11.84RR488 pKa = 11.84RR489 pKa = 11.84APRR492 pKa = 11.84ARR494 pKa = 11.84RR495 pKa = 11.84RR496 pKa = 11.84AGRR499 pKa = 11.84RR500 pKa = 11.84RR501 pKa = 11.84RR502 pKa = 11.84ASARR506 pKa = 11.84RR507 pKa = 11.84TGPRR511 pKa = 11.84GAPRR515 pKa = 11.84RR516 pKa = 11.84PGTRR520 pKa = 11.84AARR523 pKa = 11.84RR524 pKa = 11.84RR525 pKa = 11.84PGRR528 pKa = 11.84PRR530 pKa = 11.84AXASAGTALTTTSFRR545 pKa = 11.84PAAPASSPRR554 pKa = 11.84NSTAPAKK561 pKa = 10.33ARR563 pKa = 11.84SRR565 pKa = 11.84VEE567 pKa = 3.83SSSTPRR573 pKa = 11.84AFSAEE578 pKa = 4.09TSGGPVQLPAQAAEE592 pKa = 4.23SGVQRR597 pKa = 11.84RR598 pKa = 11.84LASAASRR605 pKa = 11.84NEE607 pKa = 3.85SPPRR611 pKa = 11.84SVVXLXGARR620 pKa = 11.84ARR622 pKa = 11.84ARR624 pKa = 11.84RR625 pKa = 11.84APRR628 pKa = 11.84RR629 pKa = 11.84AAARR633 pKa = 11.84RR634 pKa = 11.84ARR636 pKa = 11.84ARR638 pKa = 11.84ARR640 pKa = 11.84RR641 pKa = 11.84PGLAPGASTRR651 pKa = 11.84RR652 pKa = 11.84RR653 pKa = 11.84GAAGPRR659 pKa = 11.84RR660 pKa = 11.84AAGRR664 pKa = 11.84KK665 pKa = 9.13AGAAPALLACSRR677 pKa = 11.84ASSGAA682 pKa = 3.27
Molecular weight: 71.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
37865 |
41 |
2772 |
479.3 |
51.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.0 ± 0.484 | 1.997 ± 0.143 |
5.145 ± 0.172 | 4.838 ± 0.208 |
3.193 ± 0.157 | 7.558 ± 0.3 |
2.469 ± 0.098 | 3.251 ± 0.175 |
1.957 ± 0.134 | 8.818 ± 0.314 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.21 ± 0.108 | 2.113 ± 0.118 |
7.656 ± 0.416 | 2.673 ± 0.124 |
9.867 ± 0.537 | 7.099 ± 0.256 |
4.944 ± 0.148 | 6.626 ± 0.231 |
1.062 ± 0.067 | 2.496 ± 0.143 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
