
Pine marten torque teno virus 1
Taxonomy: Viruses; Anelloviridae; Thetatorquevirus; Torque teno mustilid virus 1
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
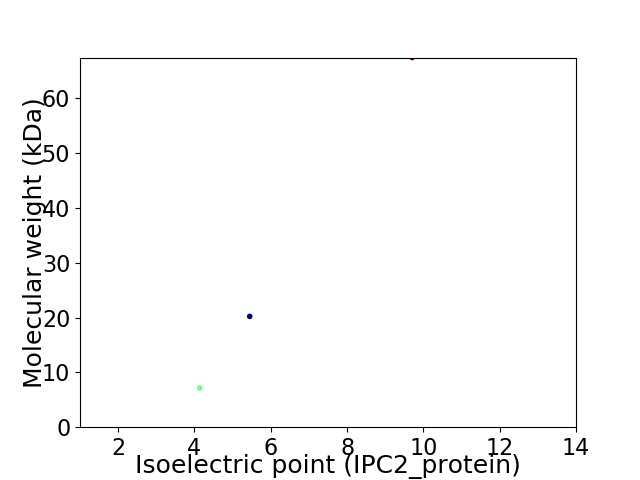
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|G9ITT1|G9ITT1_9VIRU Capsid protein OS=Pine marten torque teno virus 1 OX=1128423 PE=3 SV=1
MM1 pKa = 7.73LDD3 pKa = 3.74KK4 pKa = 10.43WLQSCADD11 pKa = 3.85THH13 pKa = 6.87SLWCKK18 pKa = 8.29CPEE21 pKa = 4.08WRR23 pKa = 11.84SHH25 pKa = 4.28VPGWRR30 pKa = 11.84GDD32 pKa = 3.67TAGDD36 pKa = 3.68GAAEE40 pKa = 4.24LPEE43 pKa = 4.37DD44 pKa = 3.72VQVAFDD50 pKa = 3.46IAFEE54 pKa = 5.17DD55 pKa = 5.22GADD58 pKa = 3.49SDD60 pKa = 3.82QPEE63 pKa = 3.94RR64 pKa = 4.02
MM1 pKa = 7.73LDD3 pKa = 3.74KK4 pKa = 10.43WLQSCADD11 pKa = 3.85THH13 pKa = 6.87SLWCKK18 pKa = 8.29CPEE21 pKa = 4.08WRR23 pKa = 11.84SHH25 pKa = 4.28VPGWRR30 pKa = 11.84GDD32 pKa = 3.67TAGDD36 pKa = 3.68GAAEE40 pKa = 4.24LPEE43 pKa = 4.37DD44 pKa = 3.72VQVAFDD50 pKa = 3.46IAFEE54 pKa = 5.17DD55 pKa = 5.22GADD58 pKa = 3.49SDD60 pKa = 3.82QPEE63 pKa = 3.94RR64 pKa = 4.02
Molecular weight: 7.15 kDa
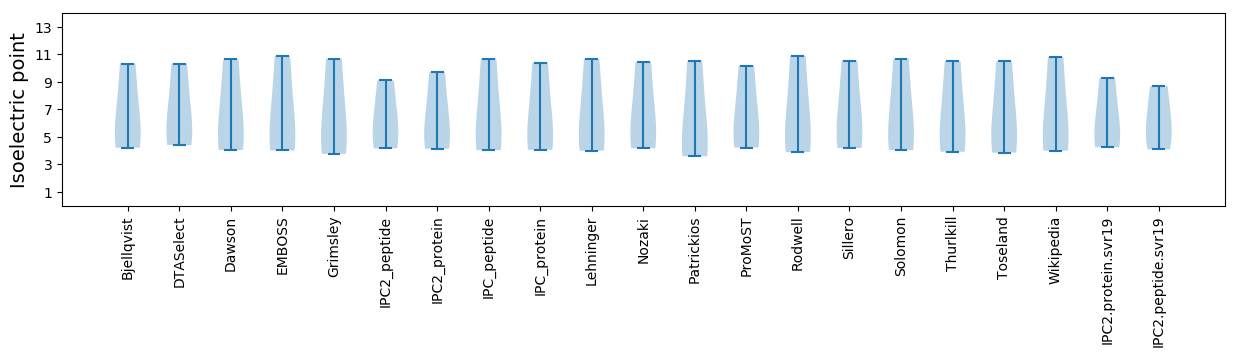
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G9ITT1|G9ITT1_9VIRU Capsid protein OS=Pine marten torque teno virus 1 OX=1128423 PE=3 SV=1
MM1 pKa = 7.37AWRR4 pKa = 11.84YY5 pKa = 6.49RR6 pKa = 11.84WRR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84APRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84AGRR20 pKa = 11.84FRR22 pKa = 11.84YY23 pKa = 9.51RR24 pKa = 11.84FRR26 pKa = 11.84RR27 pKa = 11.84WRR29 pKa = 11.84RR30 pKa = 11.84LRR32 pKa = 11.84PARR35 pKa = 11.84KK36 pKa = 7.59VRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84YY43 pKa = 9.22RR44 pKa = 11.84KK45 pKa = 8.93RR46 pKa = 11.84HH47 pKa = 4.97FKK49 pKa = 10.42KK50 pKa = 10.38KK51 pKa = 10.19AIKK54 pKa = 8.99QWQPSHH60 pKa = 5.66VARR63 pKa = 11.84CIIRR67 pKa = 11.84GWHH70 pKa = 5.42IGFWGNKK77 pKa = 9.04ANLSYY82 pKa = 10.61PFQEE86 pKa = 3.87WTRR89 pKa = 11.84PTKK92 pKa = 10.44EE93 pKa = 3.81GEE95 pKa = 3.91AGFFTIRR102 pKa = 11.84EE103 pKa = 4.37GGASLWHH110 pKa = 6.38FNLSHH115 pKa = 7.66FYY117 pKa = 11.07RR118 pKa = 11.84EE119 pKa = 4.04FTFGRR124 pKa = 11.84NTWSRR129 pKa = 11.84SNEE132 pKa = 3.85GFDD135 pKa = 3.16LATYY139 pKa = 10.22HH140 pKa = 5.1GTKK143 pKa = 10.39LIFYY147 pKa = 6.96PHH149 pKa = 7.52PLHH152 pKa = 7.27NYY154 pKa = 8.03IVWWWRR160 pKa = 11.84DD161 pKa = 3.45FGDD164 pKa = 3.81VSAAQFKK171 pKa = 10.34HH172 pKa = 5.0SHH174 pKa = 5.81PAKK177 pKa = 10.45ALLEE181 pKa = 4.33RR182 pKa = 11.84NHH184 pKa = 6.06FVIKK188 pKa = 10.34SIPQGGRR195 pKa = 11.84KK196 pKa = 7.8KK197 pKa = 7.35TTIFIPPPTTIQTGWSVMKK216 pKa = 10.38KK217 pKa = 6.96WANVNLFRR225 pKa = 11.84LGMTMINTGNPLLHH239 pKa = 7.19KK240 pKa = 10.53GRR242 pKa = 11.84LLAKK246 pKa = 10.27VPIGTYY252 pKa = 9.08WSTGNYY258 pKa = 9.76RR259 pKa = 11.84YY260 pKa = 10.14FEE262 pKa = 4.61DD263 pKa = 3.89RR264 pKa = 11.84LPVEE268 pKa = 4.48TLPEE272 pKa = 4.2ASTNIAYY279 pKa = 9.47YY280 pKa = 9.74SWWWDD285 pKa = 3.23DD286 pKa = 3.61SQDD289 pKa = 3.36NKK291 pKa = 10.53IAIRR295 pKa = 11.84DD296 pKa = 3.88NEE298 pKa = 4.0NSNDD302 pKa = 3.53GEE304 pKa = 4.82TIFHH308 pKa = 6.66LKK310 pKa = 8.86IPYY313 pKa = 8.56WLEE316 pKa = 3.46FFGAGDD322 pKa = 3.51QYY324 pKa = 11.31IEE326 pKa = 4.11QTKK329 pKa = 10.02KK330 pKa = 10.84VYY332 pKa = 9.49IWWYY336 pKa = 10.36RR337 pKa = 11.84EE338 pKa = 3.61DD339 pKa = 4.32ASIIDD344 pKa = 3.71YY345 pKa = 10.84HH346 pKa = 8.1SITSEE351 pKa = 4.87HH352 pKa = 6.13RR353 pKa = 11.84KK354 pKa = 8.63QWIKK358 pKa = 10.88LGNSSQGSISTALLAWSNIGRR379 pKa = 11.84HH380 pKa = 5.66GPFMMAEE387 pKa = 4.06EE388 pKa = 5.42DD389 pKa = 3.38ISNNSYY395 pKa = 11.57SFFFKK400 pKa = 10.85YY401 pKa = 9.91KK402 pKa = 10.58SYY404 pKa = 9.02WRR406 pKa = 11.84WGGVHH411 pKa = 6.18PTPATNNDD419 pKa = 3.71PVTVSPNSDD428 pKa = 3.44DD429 pKa = 3.66FAEE432 pKa = 4.33VQVRR436 pKa = 11.84NPLRR440 pKa = 11.84VGRR443 pKa = 11.84AALHH447 pKa = 6.29PWDD450 pKa = 5.3LDD452 pKa = 3.38PDD454 pKa = 4.26GFITTAKK461 pKa = 9.63FRR463 pKa = 11.84DD464 pKa = 3.66MLGFSPSAITGLPPPIPQRR483 pKa = 11.84RR484 pKa = 11.84AKK486 pKa = 10.4RR487 pKa = 11.84SLSPSSGEE495 pKa = 4.1SEE497 pKa = 4.24EE498 pKa = 4.36EE499 pKa = 4.36GPSVRR504 pKa = 11.84RR505 pKa = 11.84RR506 pKa = 11.84RR507 pKa = 11.84SGDD510 pKa = 3.22GEE512 pKa = 4.27LEE514 pKa = 3.94QNSFYY519 pKa = 10.99FEE521 pKa = 4.62EE522 pKa = 4.85DD523 pKa = 3.28TLPPYY528 pKa = 8.99PTFPPGEE535 pKa = 4.04TAEE538 pKa = 4.15EE539 pKa = 4.43KK540 pKa = 10.38IRR542 pKa = 11.84ALVEE546 pKa = 3.42QLRR549 pKa = 11.84RR550 pKa = 11.84DD551 pKa = 3.26RR552 pKa = 11.84DD553 pKa = 3.24KK554 pKa = 11.11RR555 pKa = 11.84RR556 pKa = 11.84KK557 pKa = 8.1LRR559 pKa = 11.84KK560 pKa = 9.4RR561 pKa = 11.84IRR563 pKa = 11.84SFLRR567 pKa = 11.84DD568 pKa = 3.11
MM1 pKa = 7.37AWRR4 pKa = 11.84YY5 pKa = 6.49RR6 pKa = 11.84WRR8 pKa = 11.84WRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84APRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84AGRR20 pKa = 11.84FRR22 pKa = 11.84YY23 pKa = 9.51RR24 pKa = 11.84FRR26 pKa = 11.84RR27 pKa = 11.84WRR29 pKa = 11.84RR30 pKa = 11.84LRR32 pKa = 11.84PARR35 pKa = 11.84KK36 pKa = 7.59VRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84YY43 pKa = 9.22RR44 pKa = 11.84KK45 pKa = 8.93RR46 pKa = 11.84HH47 pKa = 4.97FKK49 pKa = 10.42KK50 pKa = 10.38KK51 pKa = 10.19AIKK54 pKa = 8.99QWQPSHH60 pKa = 5.66VARR63 pKa = 11.84CIIRR67 pKa = 11.84GWHH70 pKa = 5.42IGFWGNKK77 pKa = 9.04ANLSYY82 pKa = 10.61PFQEE86 pKa = 3.87WTRR89 pKa = 11.84PTKK92 pKa = 10.44EE93 pKa = 3.81GEE95 pKa = 3.91AGFFTIRR102 pKa = 11.84EE103 pKa = 4.37GGASLWHH110 pKa = 6.38FNLSHH115 pKa = 7.66FYY117 pKa = 11.07RR118 pKa = 11.84EE119 pKa = 4.04FTFGRR124 pKa = 11.84NTWSRR129 pKa = 11.84SNEE132 pKa = 3.85GFDD135 pKa = 3.16LATYY139 pKa = 10.22HH140 pKa = 5.1GTKK143 pKa = 10.39LIFYY147 pKa = 6.96PHH149 pKa = 7.52PLHH152 pKa = 7.27NYY154 pKa = 8.03IVWWWRR160 pKa = 11.84DD161 pKa = 3.45FGDD164 pKa = 3.81VSAAQFKK171 pKa = 10.34HH172 pKa = 5.0SHH174 pKa = 5.81PAKK177 pKa = 10.45ALLEE181 pKa = 4.33RR182 pKa = 11.84NHH184 pKa = 6.06FVIKK188 pKa = 10.34SIPQGGRR195 pKa = 11.84KK196 pKa = 7.8KK197 pKa = 7.35TTIFIPPPTTIQTGWSVMKK216 pKa = 10.38KK217 pKa = 6.96WANVNLFRR225 pKa = 11.84LGMTMINTGNPLLHH239 pKa = 7.19KK240 pKa = 10.53GRR242 pKa = 11.84LLAKK246 pKa = 10.27VPIGTYY252 pKa = 9.08WSTGNYY258 pKa = 9.76RR259 pKa = 11.84YY260 pKa = 10.14FEE262 pKa = 4.61DD263 pKa = 3.89RR264 pKa = 11.84LPVEE268 pKa = 4.48TLPEE272 pKa = 4.2ASTNIAYY279 pKa = 9.47YY280 pKa = 9.74SWWWDD285 pKa = 3.23DD286 pKa = 3.61SQDD289 pKa = 3.36NKK291 pKa = 10.53IAIRR295 pKa = 11.84DD296 pKa = 3.88NEE298 pKa = 4.0NSNDD302 pKa = 3.53GEE304 pKa = 4.82TIFHH308 pKa = 6.66LKK310 pKa = 8.86IPYY313 pKa = 8.56WLEE316 pKa = 3.46FFGAGDD322 pKa = 3.51QYY324 pKa = 11.31IEE326 pKa = 4.11QTKK329 pKa = 10.02KK330 pKa = 10.84VYY332 pKa = 9.49IWWYY336 pKa = 10.36RR337 pKa = 11.84EE338 pKa = 3.61DD339 pKa = 4.32ASIIDD344 pKa = 3.71YY345 pKa = 10.84HH346 pKa = 8.1SITSEE351 pKa = 4.87HH352 pKa = 6.13RR353 pKa = 11.84KK354 pKa = 8.63QWIKK358 pKa = 10.88LGNSSQGSISTALLAWSNIGRR379 pKa = 11.84HH380 pKa = 5.66GPFMMAEE387 pKa = 4.06EE388 pKa = 5.42DD389 pKa = 3.38ISNNSYY395 pKa = 11.57SFFFKK400 pKa = 10.85YY401 pKa = 9.91KK402 pKa = 10.58SYY404 pKa = 9.02WRR406 pKa = 11.84WGGVHH411 pKa = 6.18PTPATNNDD419 pKa = 3.71PVTVSPNSDD428 pKa = 3.44DD429 pKa = 3.66FAEE432 pKa = 4.33VQVRR436 pKa = 11.84NPLRR440 pKa = 11.84VGRR443 pKa = 11.84AALHH447 pKa = 6.29PWDD450 pKa = 5.3LDD452 pKa = 3.38PDD454 pKa = 4.26GFITTAKK461 pKa = 9.63FRR463 pKa = 11.84DD464 pKa = 3.66MLGFSPSAITGLPPPIPQRR483 pKa = 11.84RR484 pKa = 11.84AKK486 pKa = 10.4RR487 pKa = 11.84SLSPSSGEE495 pKa = 4.1SEE497 pKa = 4.24EE498 pKa = 4.36EE499 pKa = 4.36GPSVRR504 pKa = 11.84RR505 pKa = 11.84RR506 pKa = 11.84RR507 pKa = 11.84SGDD510 pKa = 3.22GEE512 pKa = 4.27LEE514 pKa = 3.94QNSFYY519 pKa = 10.99FEE521 pKa = 4.62EE522 pKa = 4.85DD523 pKa = 3.28TLPPYY528 pKa = 8.99PTFPPGEE535 pKa = 4.04TAEE538 pKa = 4.15EE539 pKa = 4.43KK540 pKa = 10.38IRR542 pKa = 11.84ALVEE546 pKa = 3.42QLRR549 pKa = 11.84RR550 pKa = 11.84DD551 pKa = 3.26RR552 pKa = 11.84DD553 pKa = 3.24KK554 pKa = 11.11RR555 pKa = 11.84RR556 pKa = 11.84KK557 pKa = 8.1LRR559 pKa = 11.84KK560 pKa = 9.4RR561 pKa = 11.84IRR563 pKa = 11.84SFLRR567 pKa = 11.84DD568 pKa = 3.11
Molecular weight: 67.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
814 |
64 |
568 |
271.3 |
31.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.125 ± 1.574 | 0.86 ± 1.069 |
5.774 ± 2.257 | 6.511 ± 1.204 |
4.791 ± 1.364 | 7.371 ± 0.792 |
2.826 ± 0.447 | 4.791 ± 1.243 |
4.914 ± 0.841 | 6.265 ± 0.339 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.351 ± 0.136 | 3.317 ± 1.484 |
7.002 ± 0.681 | 3.808 ± 1.353 |
10.811 ± 1.533 | 7.002 ± 0.187 |
4.914 ± 0.67 | 3.194 ± 0.382 |
4.423 ± 0.718 | 2.948 ± 1.3 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
