
Ahniella affigens
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Rhodanobacteraceae; Ahniella
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
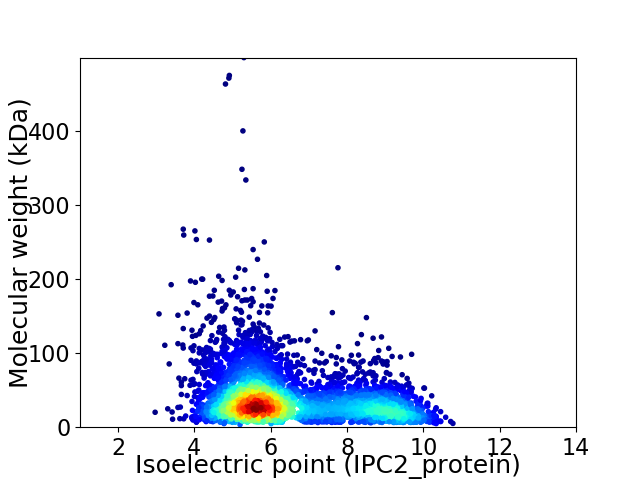
Virtual 2D-PAGE plot for 4694 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1PPF9|A0A2P1PPF9_9GAMM GspH domain-containing protein OS=Ahniella affigens OX=2021234 GN=C7S18_05660 PE=4 SV=1
MM1 pKa = 7.21MRR3 pKa = 11.84RR4 pKa = 11.84ADD6 pKa = 4.08ADD8 pKa = 3.53QCPVPSGLTKK18 pKa = 10.67LRR20 pKa = 11.84TLLWIGVCGLSACAHH35 pKa = 5.47TPKK38 pKa = 10.2PDD40 pKa = 3.63ADD42 pKa = 3.67GAGANSAVRR51 pKa = 11.84SPAPVTATADD61 pKa = 3.38LGQPMSVQTEE71 pKa = 4.03ALAVQQSNGSAQYY84 pKa = 10.69QRR86 pKa = 11.84LFPARR91 pKa = 11.84TDD93 pKa = 3.42TQPQFQVPHH102 pKa = 6.29ARR104 pKa = 11.84WPVLPGTRR112 pKa = 11.84GRR114 pKa = 11.84GNASAPEE121 pKa = 3.96RR122 pKa = 11.84VQGGGAAVAEE132 pKa = 4.46ATAGSPSDD140 pKa = 3.55RR141 pKa = 11.84PTAVGDD147 pKa = 3.33AAKK150 pKa = 9.91IVGSIAPKK158 pKa = 9.76FDD160 pKa = 4.97SIAFDD165 pKa = 5.39DD166 pKa = 4.98NGTLNDD172 pKa = 3.68GFRR175 pKa = 11.84FIPADD180 pKa = 3.22AHH182 pKa = 6.3AAAGPNHH189 pKa = 5.69VVAVSNVTLKK199 pKa = 10.05IHH201 pKa = 6.61RR202 pKa = 11.84KK203 pKa = 9.58SDD205 pKa = 3.39GQVQLQTGLQDD216 pKa = 3.62FFAGLGSQRR225 pKa = 11.84PSTYY229 pKa = 10.07TFDD232 pKa = 3.59PRR234 pKa = 11.84VLFDD238 pKa = 3.63PFEE241 pKa = 4.07NRR243 pKa = 11.84FVVLTLEE250 pKa = 4.46VLSQANGDD258 pKa = 3.76ATDD261 pKa = 3.49SSFLFVAVSDD271 pKa = 4.23DD272 pKa = 3.8ADD274 pKa = 4.06PNGVWRR280 pKa = 11.84MARR283 pKa = 11.84INTKK287 pKa = 8.92LTISSTPGWLDD298 pKa = 3.35YY299 pKa = 10.99PGFGIDD305 pKa = 3.83EE306 pKa = 4.7DD307 pKa = 4.03ALYY310 pKa = 9.39VTGNYY315 pKa = 9.85FDD317 pKa = 3.98FTVGNYY323 pKa = 9.91LDD325 pKa = 3.57SRR327 pKa = 11.84LWILNKK333 pKa = 10.08GVSGGLYY340 pKa = 10.21SGGTVVARR348 pKa = 11.84QFDD351 pKa = 4.52PEE353 pKa = 4.1PTLADD358 pKa = 3.72SAPVMPARR366 pKa = 11.84MLSAGPGTVGTWLAAYY382 pKa = 10.23SGFATTGGVEE392 pKa = 4.25QINTIRR398 pKa = 11.84IDD400 pKa = 3.61NPLNTSPTFTSALEE414 pKa = 3.99NVADD418 pKa = 3.83IDD420 pKa = 4.02QSFTVDD426 pKa = 4.73FPNAPQPGSVAGLDD440 pKa = 3.43PGDD443 pKa = 3.65RR444 pKa = 11.84RR445 pKa = 11.84MLDD448 pKa = 3.59VVWHH452 pKa = 5.75NNRR455 pKa = 11.84LSMVFNVLPPASQSNANQNTAYY477 pKa = 9.54WLQLDD482 pKa = 3.92TSVLNNLSVAQRR494 pKa = 11.84GQIGGEE500 pKa = 3.88DD501 pKa = 3.8LGSSVHH507 pKa = 5.56TFYY510 pKa = 10.9PSIATNQFGHH520 pKa = 6.92LAISFAASSTNLNPGAYY537 pKa = 9.16VVTRR541 pKa = 11.84RR542 pKa = 11.84PTDD545 pKa = 3.35SNNTVSSSQTLRR557 pKa = 11.84AGLDD561 pKa = 3.13TYY563 pKa = 11.5LRR565 pKa = 11.84TFGGNRR571 pKa = 11.84NRR573 pKa = 11.84WGDD576 pKa = 3.57YY577 pKa = 9.46SATMVDD583 pKa = 3.72PTDD586 pKa = 3.26QCFWSFNLWSNTNGTVISGEE606 pKa = 4.01DD607 pKa = 3.86GRR609 pKa = 11.84WSTTAGRR616 pKa = 11.84LCTCEE621 pKa = 4.16GTEE624 pKa = 4.17STGDD628 pKa = 3.21GDD630 pKa = 4.65FDD632 pKa = 3.8GWCNSLDD639 pKa = 3.72NCDD642 pKa = 4.88LISNRR647 pKa = 11.84DD648 pKa = 3.56QLDD651 pKa = 3.77PDD653 pKa = 3.93SDD655 pKa = 4.47TVGTACDD662 pKa = 3.63NCPNAANTNQADD674 pKa = 3.75GDD676 pKa = 3.95NDD678 pKa = 3.65QVGTVCDD685 pKa = 3.63NCSTVANTNQANADD699 pKa = 3.12ADD701 pKa = 3.78ARR703 pKa = 11.84GDD705 pKa = 3.61VCDD708 pKa = 3.61NCVNVANNDD717 pKa = 3.9QINSDD722 pKa = 3.68ADD724 pKa = 4.01SVGDD728 pKa = 3.5ACDD731 pKa = 3.31NCRR734 pKa = 11.84LISNQNQSNLDD745 pKa = 3.5FDD747 pKa = 4.36QLGDD751 pKa = 3.73ACDD754 pKa = 3.75NCPFAANNDD763 pKa = 3.83QLNSDD768 pKa = 3.73TDD770 pKa = 4.07GLGNACDD777 pKa = 3.74NCPLVTNQSQINSDD791 pKa = 3.51SDD793 pKa = 4.1LLGDD797 pKa = 4.44ACDD800 pKa = 3.71NCISVANASQTNSDD814 pKa = 3.62SDD816 pKa = 4.31LLGDD820 pKa = 4.56ACDD823 pKa = 3.79NCDD826 pKa = 5.29LISNPDD832 pKa = 3.35QADD835 pKa = 3.36GDD837 pKa = 4.06GDD839 pKa = 4.44LVGTACDD846 pKa = 3.47NCPAAGNTNQTNSDD860 pKa = 3.98GDD862 pKa = 3.93PLGDD866 pKa = 3.46VCDD869 pKa = 3.86NCVTVTNPDD878 pKa = 3.44QLNTDD883 pKa = 3.49MDD885 pKa = 4.62SLGDD889 pKa = 3.76ACDD892 pKa = 4.26GDD894 pKa = 4.6DD895 pKa = 5.93DD896 pKa = 5.13NDD898 pKa = 3.93AVPDD902 pKa = 4.0GTDD905 pKa = 3.19NCPLVPNLNQTNTDD919 pKa = 3.45GANDD923 pKa = 4.3GGDD926 pKa = 3.39ACDD929 pKa = 4.86PDD931 pKa = 5.27DD932 pKa = 6.77DD933 pKa = 5.29NDD935 pKa = 4.11TVLDD939 pKa = 4.31GVDD942 pKa = 3.54NCPLIVNLNQANADD956 pKa = 3.43GDD958 pKa = 4.22ALGDD962 pKa = 3.67VCDD965 pKa = 5.31PDD967 pKa = 5.26DD968 pKa = 6.29DD969 pKa = 5.07NDD971 pKa = 3.97GFQDD975 pKa = 4.5ASDD978 pKa = 3.82NCPLVSNVDD987 pKa = 3.6QLDD990 pKa = 3.81DD991 pKa = 5.48DD992 pKa = 4.82GDD994 pKa = 3.89LRR996 pKa = 11.84GNVCDD1001 pKa = 3.74NCPAISNADD1010 pKa = 3.4QADD1013 pKa = 3.76RR1014 pKa = 11.84DD1015 pKa = 3.49LDD1017 pKa = 3.61QRR1019 pKa = 11.84GNVCDD1024 pKa = 3.82NCPFAANPSQQDD1036 pKa = 3.35DD1037 pKa = 4.05DD1038 pKa = 5.37GDD1040 pKa = 4.68LIGNVCDD1047 pKa = 4.5FCPNEE1052 pKa = 4.45FSNQCYY1058 pKa = 9.8VFGDD1062 pKa = 4.47GYY1064 pKa = 11.1EE1065 pKa = 4.56DD1066 pKa = 4.64PLTLL1070 pKa = 4.98
MM1 pKa = 7.21MRR3 pKa = 11.84RR4 pKa = 11.84ADD6 pKa = 4.08ADD8 pKa = 3.53QCPVPSGLTKK18 pKa = 10.67LRR20 pKa = 11.84TLLWIGVCGLSACAHH35 pKa = 5.47TPKK38 pKa = 10.2PDD40 pKa = 3.63ADD42 pKa = 3.67GAGANSAVRR51 pKa = 11.84SPAPVTATADD61 pKa = 3.38LGQPMSVQTEE71 pKa = 4.03ALAVQQSNGSAQYY84 pKa = 10.69QRR86 pKa = 11.84LFPARR91 pKa = 11.84TDD93 pKa = 3.42TQPQFQVPHH102 pKa = 6.29ARR104 pKa = 11.84WPVLPGTRR112 pKa = 11.84GRR114 pKa = 11.84GNASAPEE121 pKa = 3.96RR122 pKa = 11.84VQGGGAAVAEE132 pKa = 4.46ATAGSPSDD140 pKa = 3.55RR141 pKa = 11.84PTAVGDD147 pKa = 3.33AAKK150 pKa = 9.91IVGSIAPKK158 pKa = 9.76FDD160 pKa = 4.97SIAFDD165 pKa = 5.39DD166 pKa = 4.98NGTLNDD172 pKa = 3.68GFRR175 pKa = 11.84FIPADD180 pKa = 3.22AHH182 pKa = 6.3AAAGPNHH189 pKa = 5.69VVAVSNVTLKK199 pKa = 10.05IHH201 pKa = 6.61RR202 pKa = 11.84KK203 pKa = 9.58SDD205 pKa = 3.39GQVQLQTGLQDD216 pKa = 3.62FFAGLGSQRR225 pKa = 11.84PSTYY229 pKa = 10.07TFDD232 pKa = 3.59PRR234 pKa = 11.84VLFDD238 pKa = 3.63PFEE241 pKa = 4.07NRR243 pKa = 11.84FVVLTLEE250 pKa = 4.46VLSQANGDD258 pKa = 3.76ATDD261 pKa = 3.49SSFLFVAVSDD271 pKa = 4.23DD272 pKa = 3.8ADD274 pKa = 4.06PNGVWRR280 pKa = 11.84MARR283 pKa = 11.84INTKK287 pKa = 8.92LTISSTPGWLDD298 pKa = 3.35YY299 pKa = 10.99PGFGIDD305 pKa = 3.83EE306 pKa = 4.7DD307 pKa = 4.03ALYY310 pKa = 9.39VTGNYY315 pKa = 9.85FDD317 pKa = 3.98FTVGNYY323 pKa = 9.91LDD325 pKa = 3.57SRR327 pKa = 11.84LWILNKK333 pKa = 10.08GVSGGLYY340 pKa = 10.21SGGTVVARR348 pKa = 11.84QFDD351 pKa = 4.52PEE353 pKa = 4.1PTLADD358 pKa = 3.72SAPVMPARR366 pKa = 11.84MLSAGPGTVGTWLAAYY382 pKa = 10.23SGFATTGGVEE392 pKa = 4.25QINTIRR398 pKa = 11.84IDD400 pKa = 3.61NPLNTSPTFTSALEE414 pKa = 3.99NVADD418 pKa = 3.83IDD420 pKa = 4.02QSFTVDD426 pKa = 4.73FPNAPQPGSVAGLDD440 pKa = 3.43PGDD443 pKa = 3.65RR444 pKa = 11.84RR445 pKa = 11.84MLDD448 pKa = 3.59VVWHH452 pKa = 5.75NNRR455 pKa = 11.84LSMVFNVLPPASQSNANQNTAYY477 pKa = 9.54WLQLDD482 pKa = 3.92TSVLNNLSVAQRR494 pKa = 11.84GQIGGEE500 pKa = 3.88DD501 pKa = 3.8LGSSVHH507 pKa = 5.56TFYY510 pKa = 10.9PSIATNQFGHH520 pKa = 6.92LAISFAASSTNLNPGAYY537 pKa = 9.16VVTRR541 pKa = 11.84RR542 pKa = 11.84PTDD545 pKa = 3.35SNNTVSSSQTLRR557 pKa = 11.84AGLDD561 pKa = 3.13TYY563 pKa = 11.5LRR565 pKa = 11.84TFGGNRR571 pKa = 11.84NRR573 pKa = 11.84WGDD576 pKa = 3.57YY577 pKa = 9.46SATMVDD583 pKa = 3.72PTDD586 pKa = 3.26QCFWSFNLWSNTNGTVISGEE606 pKa = 4.01DD607 pKa = 3.86GRR609 pKa = 11.84WSTTAGRR616 pKa = 11.84LCTCEE621 pKa = 4.16GTEE624 pKa = 4.17STGDD628 pKa = 3.21GDD630 pKa = 4.65FDD632 pKa = 3.8GWCNSLDD639 pKa = 3.72NCDD642 pKa = 4.88LISNRR647 pKa = 11.84DD648 pKa = 3.56QLDD651 pKa = 3.77PDD653 pKa = 3.93SDD655 pKa = 4.47TVGTACDD662 pKa = 3.63NCPNAANTNQADD674 pKa = 3.75GDD676 pKa = 3.95NDD678 pKa = 3.65QVGTVCDD685 pKa = 3.63NCSTVANTNQANADD699 pKa = 3.12ADD701 pKa = 3.78ARR703 pKa = 11.84GDD705 pKa = 3.61VCDD708 pKa = 3.61NCVNVANNDD717 pKa = 3.9QINSDD722 pKa = 3.68ADD724 pKa = 4.01SVGDD728 pKa = 3.5ACDD731 pKa = 3.31NCRR734 pKa = 11.84LISNQNQSNLDD745 pKa = 3.5FDD747 pKa = 4.36QLGDD751 pKa = 3.73ACDD754 pKa = 3.75NCPFAANNDD763 pKa = 3.83QLNSDD768 pKa = 3.73TDD770 pKa = 4.07GLGNACDD777 pKa = 3.74NCPLVTNQSQINSDD791 pKa = 3.51SDD793 pKa = 4.1LLGDD797 pKa = 4.44ACDD800 pKa = 3.71NCISVANASQTNSDD814 pKa = 3.62SDD816 pKa = 4.31LLGDD820 pKa = 4.56ACDD823 pKa = 3.79NCDD826 pKa = 5.29LISNPDD832 pKa = 3.35QADD835 pKa = 3.36GDD837 pKa = 4.06GDD839 pKa = 4.44LVGTACDD846 pKa = 3.47NCPAAGNTNQTNSDD860 pKa = 3.98GDD862 pKa = 3.93PLGDD866 pKa = 3.46VCDD869 pKa = 3.86NCVTVTNPDD878 pKa = 3.44QLNTDD883 pKa = 3.49MDD885 pKa = 4.62SLGDD889 pKa = 3.76ACDD892 pKa = 4.26GDD894 pKa = 4.6DD895 pKa = 5.93DD896 pKa = 5.13NDD898 pKa = 3.93AVPDD902 pKa = 4.0GTDD905 pKa = 3.19NCPLVPNLNQTNTDD919 pKa = 3.45GANDD923 pKa = 4.3GGDD926 pKa = 3.39ACDD929 pKa = 4.86PDD931 pKa = 5.27DD932 pKa = 6.77DD933 pKa = 5.29NDD935 pKa = 4.11TVLDD939 pKa = 4.31GVDD942 pKa = 3.54NCPLIVNLNQANADD956 pKa = 3.43GDD958 pKa = 4.22ALGDD962 pKa = 3.67VCDD965 pKa = 5.31PDD967 pKa = 5.26DD968 pKa = 6.29DD969 pKa = 5.07NDD971 pKa = 3.97GFQDD975 pKa = 4.5ASDD978 pKa = 3.82NCPLVSNVDD987 pKa = 3.6QLDD990 pKa = 3.81DD991 pKa = 5.48DD992 pKa = 4.82GDD994 pKa = 3.89LRR996 pKa = 11.84GNVCDD1001 pKa = 3.74NCPAISNADD1010 pKa = 3.4QADD1013 pKa = 3.76RR1014 pKa = 11.84DD1015 pKa = 3.49LDD1017 pKa = 3.61QRR1019 pKa = 11.84GNVCDD1024 pKa = 3.82NCPFAANPSQQDD1036 pKa = 3.35DD1037 pKa = 4.05DD1038 pKa = 5.37GDD1040 pKa = 4.68LIGNVCDD1047 pKa = 4.5FCPNEE1052 pKa = 4.45FSNQCYY1058 pKa = 9.8VFGDD1062 pKa = 4.47GYY1064 pKa = 11.1EE1065 pKa = 4.56DD1066 pKa = 4.64PLTLL1070 pKa = 4.98
Molecular weight: 112.74 kDa
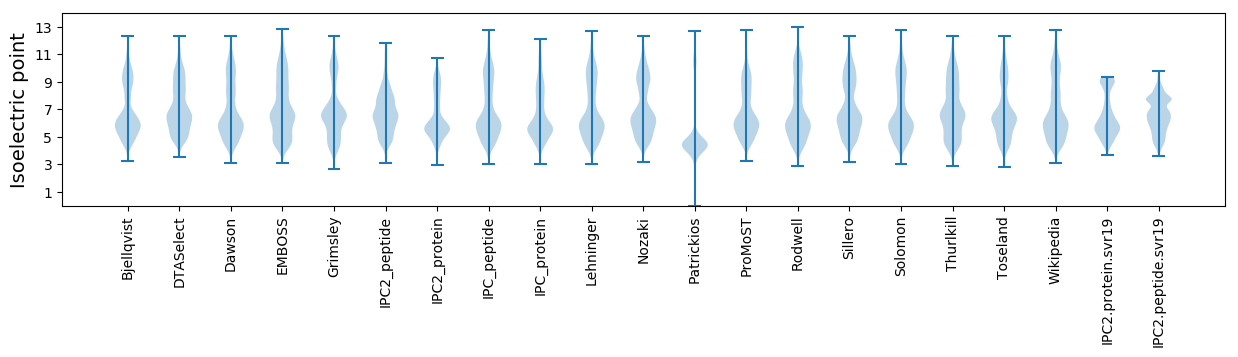
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1PLR2|A0A2P1PLR2_9GAMM Alpha/beta hydrolase OS=Ahniella affigens OX=2021234 GN=C7S18_00605 PE=4 SV=1
MM1 pKa = 7.49GKK3 pKa = 7.8TRR5 pKa = 11.84AEE7 pKa = 4.25LNTPSDD13 pKa = 3.73LATAITRR20 pKa = 11.84CAGHH24 pKa = 5.82VAALVSKK31 pKa = 10.39EE32 pKa = 3.57LDD34 pKa = 3.31ALVRR38 pKa = 11.84TARR41 pKa = 11.84TLQEE45 pKa = 3.91QYY47 pKa = 10.37EE48 pKa = 4.39SRR50 pKa = 11.84RR51 pKa = 11.84TAEE54 pKa = 3.87EE55 pKa = 3.83TRR57 pKa = 11.84WQHH60 pKa = 4.65MVLAIKK66 pKa = 9.03TGPSLVLQWSRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84IRR82 pKa = 11.84RR83 pKa = 11.84TATGRR88 pKa = 11.84AWWWDD93 pKa = 3.31TMPYY97 pKa = 9.53DD98 pKa = 4.83AKK100 pKa = 10.69RR101 pKa = 11.84RR102 pKa = 11.84CCPIRR107 pKa = 11.84ALTANQPPEE116 pKa = 3.64IAALIISTEE125 pKa = 4.02SAATEE130 pKa = 4.0IRR132 pKa = 11.84KK133 pKa = 8.54NWQLVTRR140 pKa = 11.84LRR142 pKa = 11.84ITATALMRR150 pKa = 11.84LGQLAPAEE158 pKa = 4.65SPWLHH163 pKa = 5.42LTGKK167 pKa = 10.1NGSTILAQDD176 pKa = 3.81HH177 pKa = 5.82MQDD180 pKa = 3.47PDD182 pKa = 3.74SFATTVLMFVAALQDD197 pKa = 3.4QLLRR201 pKa = 11.84NGDD204 pKa = 3.81ALSQYY209 pKa = 10.51VEE211 pKa = 4.18NNRR214 pKa = 11.84SNKK217 pKa = 9.58GYY219 pKa = 10.19RR220 pKa = 11.84LRR222 pKa = 11.84SNRR225 pKa = 11.84RR226 pKa = 11.84GNSLDD231 pKa = 3.49LAWYY235 pKa = 8.62HH236 pKa = 6.35GPAASRR242 pKa = 11.84TTVTRR247 pKa = 11.84RR248 pKa = 11.84KK249 pKa = 9.22NANITRR255 pKa = 11.84LIATNSEE262 pKa = 3.97DD263 pKa = 3.34ANLIRR268 pKa = 11.84EE269 pKa = 4.47CEE271 pKa = 3.78AAATVIRR278 pKa = 11.84RR279 pKa = 11.84AWAMCGQLIARR290 pKa = 11.84AKK292 pKa = 10.23EE293 pKa = 3.8IEE295 pKa = 4.41KK296 pKa = 9.12LTKK299 pKa = 10.12ACAPRR304 pKa = 11.84KK305 pKa = 9.56LGEE308 pKa = 3.83IAMGGLGGNPEE319 pKa = 4.21APPRR323 pKa = 11.84SSKK326 pKa = 11.07
MM1 pKa = 7.49GKK3 pKa = 7.8TRR5 pKa = 11.84AEE7 pKa = 4.25LNTPSDD13 pKa = 3.73LATAITRR20 pKa = 11.84CAGHH24 pKa = 5.82VAALVSKK31 pKa = 10.39EE32 pKa = 3.57LDD34 pKa = 3.31ALVRR38 pKa = 11.84TARR41 pKa = 11.84TLQEE45 pKa = 3.91QYY47 pKa = 10.37EE48 pKa = 4.39SRR50 pKa = 11.84RR51 pKa = 11.84TAEE54 pKa = 3.87EE55 pKa = 3.83TRR57 pKa = 11.84WQHH60 pKa = 4.65MVLAIKK66 pKa = 9.03TGPSLVLQWSRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84IRR82 pKa = 11.84RR83 pKa = 11.84TATGRR88 pKa = 11.84AWWWDD93 pKa = 3.31TMPYY97 pKa = 9.53DD98 pKa = 4.83AKK100 pKa = 10.69RR101 pKa = 11.84RR102 pKa = 11.84CCPIRR107 pKa = 11.84ALTANQPPEE116 pKa = 3.64IAALIISTEE125 pKa = 4.02SAATEE130 pKa = 4.0IRR132 pKa = 11.84KK133 pKa = 8.54NWQLVTRR140 pKa = 11.84LRR142 pKa = 11.84ITATALMRR150 pKa = 11.84LGQLAPAEE158 pKa = 4.65SPWLHH163 pKa = 5.42LTGKK167 pKa = 10.1NGSTILAQDD176 pKa = 3.81HH177 pKa = 5.82MQDD180 pKa = 3.47PDD182 pKa = 3.74SFATTVLMFVAALQDD197 pKa = 3.4QLLRR201 pKa = 11.84NGDD204 pKa = 3.81ALSQYY209 pKa = 10.51VEE211 pKa = 4.18NNRR214 pKa = 11.84SNKK217 pKa = 9.58GYY219 pKa = 10.19RR220 pKa = 11.84LRR222 pKa = 11.84SNRR225 pKa = 11.84RR226 pKa = 11.84GNSLDD231 pKa = 3.49LAWYY235 pKa = 8.62HH236 pKa = 6.35GPAASRR242 pKa = 11.84TTVTRR247 pKa = 11.84RR248 pKa = 11.84KK249 pKa = 9.22NANITRR255 pKa = 11.84LIATNSEE262 pKa = 3.97DD263 pKa = 3.34ANLIRR268 pKa = 11.84EE269 pKa = 4.47CEE271 pKa = 3.78AAATVIRR278 pKa = 11.84RR279 pKa = 11.84AWAMCGQLIARR290 pKa = 11.84AKK292 pKa = 10.23EE293 pKa = 3.8IEE295 pKa = 4.41KK296 pKa = 9.12LTKK299 pKa = 10.12ACAPRR304 pKa = 11.84KK305 pKa = 9.56LGEE308 pKa = 3.83IAMGGLGGNPEE319 pKa = 4.21APPRR323 pKa = 11.84SSKK326 pKa = 11.07
Molecular weight: 36.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1753483 |
30 |
4550 |
373.6 |
40.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.987 ± 0.041 | 0.969 ± 0.011 |
5.824 ± 0.028 | 5.114 ± 0.034 |
3.67 ± 0.02 | 7.96 ± 0.035 |
2.122 ± 0.019 | 4.568 ± 0.022 |
2.794 ± 0.03 | 10.668 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.057 ± 0.016 | 3.2 ± 0.033 |
5.264 ± 0.033 | 4.194 ± 0.021 |
7.005 ± 0.044 | 6.186 ± 0.037 |
5.504 ± 0.05 | 7.128 ± 0.031 |
1.536 ± 0.018 | 2.251 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
