
Bellinger River virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Tornidovirineae; Tobaniviridae; Serpentovirinae; Pregotovirus; Snaturtovirus; Berisnavirus 1
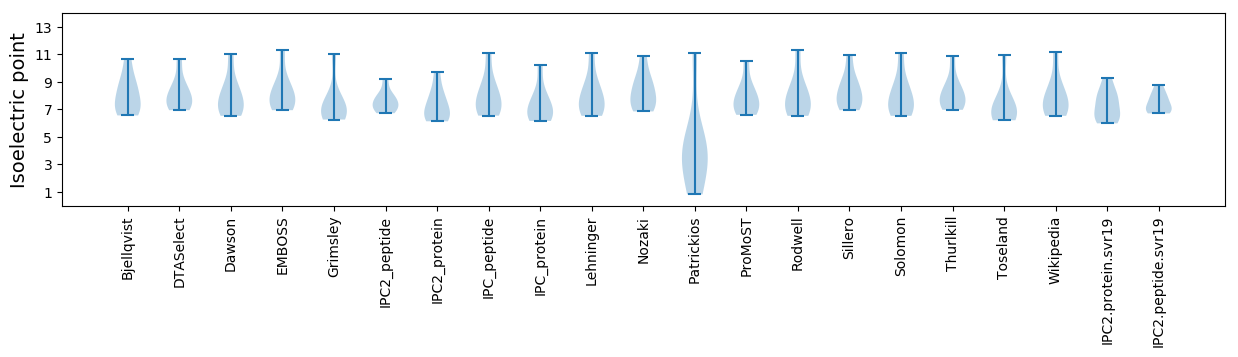
Average proteome isoelectric point is 7.31
Get precalculated fractions of proteins
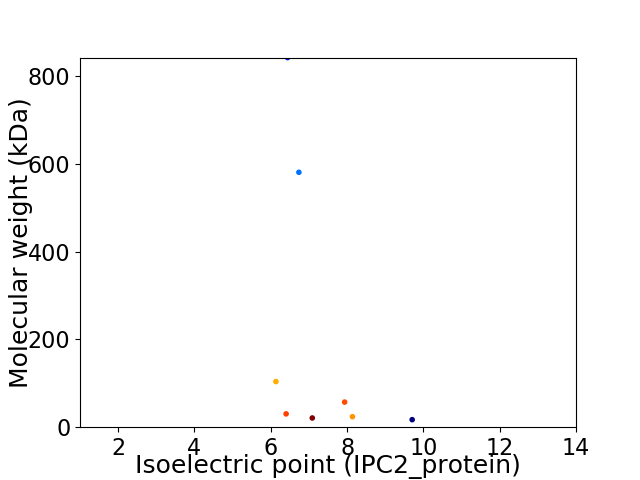
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346I7I9|A0A346I7I9_9NIDO Minor membrane protein 1 OS=Bellinger River virus OX=2301728 GN=MM1 PE=4 SV=1
MM1 pKa = 7.31FLVLLSILATCEE13 pKa = 4.39CQSHH17 pKa = 7.03RR18 pKa = 11.84YY19 pKa = 9.48DD20 pKa = 3.07NWDD23 pKa = 3.58KK24 pKa = 11.58VFTLDD29 pKa = 5.2NNIQNIISQKK39 pKa = 10.0QFDD42 pKa = 4.4NFLEE46 pKa = 4.3QPFLINSISNDD57 pKa = 3.34VKK59 pKa = 8.71PTEE62 pKa = 3.78IVEE65 pKa = 4.29YY66 pKa = 9.33LTNNQLTQALPNKK79 pKa = 9.52LPHH82 pKa = 6.33PVISSVKK89 pKa = 10.15IIDD92 pKa = 4.0KK93 pKa = 8.68QTTACNSQLINCDD106 pKa = 3.43TKK108 pKa = 11.36VKK110 pKa = 9.86TLTNLNGNCFFFNPSSISNRR130 pKa = 11.84SCYY133 pKa = 9.66KK134 pKa = 9.4NKK136 pKa = 9.2IWRR139 pKa = 11.84VRR141 pKa = 11.84NTNDD145 pKa = 4.14NSSTCDD151 pKa = 3.2NVYY154 pKa = 11.12APIGVKK160 pKa = 9.8TITQDD165 pKa = 3.13GKK167 pKa = 10.26QVPLYY172 pKa = 10.07YY173 pKa = 10.15HH174 pKa = 7.73DD175 pKa = 5.36ILPQLYY181 pKa = 9.13EE182 pKa = 3.87PGRR185 pKa = 11.84IVNDD189 pKa = 3.14NRR191 pKa = 11.84EE192 pKa = 4.15CCIFKK197 pKa = 10.0TPSVEE202 pKa = 4.32CKK204 pKa = 10.28SKK206 pKa = 11.07AVLAGGKK213 pKa = 7.1ITNYY217 pKa = 9.71INTFHH222 pKa = 6.36EE223 pKa = 4.75TIIQCPCAQKK233 pKa = 10.7CVIFINSQIDD243 pKa = 3.55VLISDD248 pKa = 5.59GILIKK253 pKa = 10.28STFEE257 pKa = 4.61LEE259 pKa = 3.88NLHH262 pKa = 6.62NIKK265 pKa = 10.26DD266 pKa = 3.64QHH268 pKa = 5.88TKK270 pKa = 10.69SILRR274 pKa = 11.84QQQPEE279 pKa = 4.1TSKK282 pKa = 11.35LEE284 pKa = 3.94RR285 pKa = 11.84QYY287 pKa = 11.83LEE289 pKa = 4.45FNNNCGDD296 pKa = 4.23SITKK300 pKa = 10.12AGCEE304 pKa = 3.84LEE306 pKa = 4.28TIEE309 pKa = 4.8FSSSIYY315 pKa = 9.31MIHH318 pKa = 6.36NAVMPNTIFKK328 pKa = 8.0TFCAVEE334 pKa = 4.09KK335 pKa = 11.02LPINDD340 pKa = 3.37QLEE343 pKa = 4.21PLNSKK348 pKa = 10.2FSVPATVQLGKK359 pKa = 10.12QRR361 pKa = 11.84YY362 pKa = 7.88IAHH365 pKa = 6.95KK366 pKa = 9.06VNPVTFQLDD375 pKa = 3.67YY376 pKa = 11.43TNDD379 pKa = 3.54PDD381 pKa = 5.41FISATTTTLPTIGSCRR397 pKa = 11.84RR398 pKa = 11.84SDD400 pKa = 3.27GYY402 pKa = 11.09AWHH405 pKa = 7.06GNFSIEE411 pKa = 4.08KK412 pKa = 8.38NEE414 pKa = 4.63ARR416 pKa = 11.84IEE418 pKa = 4.32DD419 pKa = 4.19CTATTLDD426 pKa = 3.95WNPVCNITKK435 pKa = 10.42LEE437 pKa = 4.13PLVKK441 pKa = 10.33VYY443 pKa = 10.78AVDD446 pKa = 3.83KK447 pKa = 10.73KK448 pKa = 9.88ITAKK452 pKa = 10.47GVNCDD457 pKa = 4.78DD458 pKa = 3.48ICPYY462 pKa = 11.04AVLCSEE468 pKa = 4.16EE469 pKa = 4.36QKK471 pKa = 10.67RR472 pKa = 11.84SPIYY476 pKa = 9.19QACLAIEE483 pKa = 4.02QDD485 pKa = 2.87ISTLLKK491 pKa = 10.2VEE493 pKa = 4.55RR494 pKa = 11.84PTSVKK499 pKa = 9.53MAALTKK505 pKa = 9.64TEE507 pKa = 4.32PLAGTIDD514 pKa = 3.62VNFLQEE520 pKa = 4.01VAKK523 pKa = 10.85VQAQRR528 pKa = 11.84FDD530 pKa = 3.74LTVEE534 pKa = 3.99TGLDD538 pKa = 3.38HH539 pKa = 6.65TQVLDD544 pKa = 4.11DD545 pKa = 4.35LFKK548 pKa = 10.88LTVALPQDD556 pKa = 3.56STFFPDD562 pKa = 3.61RR563 pKa = 11.84SQAAIDD569 pKa = 4.32LGLFQNAMFNSKK581 pKa = 10.72ADD583 pKa = 3.72MDD585 pKa = 4.31HH586 pKa = 6.34VAAFPWLAGWRR597 pKa = 11.84FGRR600 pKa = 11.84QINGLSYY607 pKa = 8.05TTSNLIRR614 pKa = 11.84SYY616 pKa = 10.77RR617 pKa = 11.84QFSNTVNKK625 pKa = 10.14NFQQVATALSAMADD639 pKa = 3.97QITMNYY645 pKa = 9.49EE646 pKa = 3.65VTTRR650 pKa = 11.84LYY652 pKa = 10.99EE653 pKa = 4.38SIQRR657 pKa = 11.84SYY659 pKa = 10.36QAYY662 pKa = 8.5EE663 pKa = 3.85NNFAKK668 pKa = 9.69IDD670 pKa = 3.65YY671 pKa = 9.74RR672 pKa = 11.84MSEE675 pKa = 4.21IEE677 pKa = 4.97FMTAKK682 pKa = 10.55LSLINQYY689 pKa = 7.41FTKK692 pKa = 10.34IATRR696 pKa = 11.84QSQLASEE703 pKa = 4.39HH704 pKa = 6.18QIFDD708 pKa = 4.03LRR710 pKa = 11.84VGSCRR715 pKa = 11.84TANPACFGGQGVYY728 pKa = 9.73LAHH731 pKa = 7.42AEE733 pKa = 4.26FEE735 pKa = 4.6TPSQKK740 pKa = 10.94LLIVNYY746 pKa = 10.22LGSTEE751 pKa = 3.85KK752 pKa = 10.65CKK754 pKa = 10.49QVFTANKK761 pKa = 8.74ICTKK765 pKa = 10.5QGTATIAPFGCAFFKK780 pKa = 10.78TSDD783 pKa = 3.47SDD785 pKa = 4.62SEE787 pKa = 4.32VLRR790 pKa = 11.84NLKK793 pKa = 10.67DD794 pKa = 3.28NGPCHH799 pKa = 6.52VPPINVQDD807 pKa = 3.97CQLPTNEE814 pKa = 4.83IIQLQLINYY823 pKa = 9.21QEE825 pKa = 4.11TLEE828 pKa = 4.24LGIRR832 pKa = 11.84QVNLTRR838 pKa = 11.84VEE840 pKa = 4.34FNQKK844 pKa = 8.74IANISTFNNNIKK856 pKa = 10.35QLVNSIDD863 pKa = 3.76KK864 pKa = 10.73VPSLGDD870 pKa = 3.35MLEE873 pKa = 4.17KK874 pKa = 9.8QTIASLNSIIAQINAWSIWDD894 pKa = 3.64IIKK897 pKa = 9.99WFLVCGVIIAFSALGISVIKK917 pKa = 10.68LVTKK921 pKa = 10.58RR922 pKa = 3.56
MM1 pKa = 7.31FLVLLSILATCEE13 pKa = 4.39CQSHH17 pKa = 7.03RR18 pKa = 11.84YY19 pKa = 9.48DD20 pKa = 3.07NWDD23 pKa = 3.58KK24 pKa = 11.58VFTLDD29 pKa = 5.2NNIQNIISQKK39 pKa = 10.0QFDD42 pKa = 4.4NFLEE46 pKa = 4.3QPFLINSISNDD57 pKa = 3.34VKK59 pKa = 8.71PTEE62 pKa = 3.78IVEE65 pKa = 4.29YY66 pKa = 9.33LTNNQLTQALPNKK79 pKa = 9.52LPHH82 pKa = 6.33PVISSVKK89 pKa = 10.15IIDD92 pKa = 4.0KK93 pKa = 8.68QTTACNSQLINCDD106 pKa = 3.43TKK108 pKa = 11.36VKK110 pKa = 9.86TLTNLNGNCFFFNPSSISNRR130 pKa = 11.84SCYY133 pKa = 9.66KK134 pKa = 9.4NKK136 pKa = 9.2IWRR139 pKa = 11.84VRR141 pKa = 11.84NTNDD145 pKa = 4.14NSSTCDD151 pKa = 3.2NVYY154 pKa = 11.12APIGVKK160 pKa = 9.8TITQDD165 pKa = 3.13GKK167 pKa = 10.26QVPLYY172 pKa = 10.07YY173 pKa = 10.15HH174 pKa = 7.73DD175 pKa = 5.36ILPQLYY181 pKa = 9.13EE182 pKa = 3.87PGRR185 pKa = 11.84IVNDD189 pKa = 3.14NRR191 pKa = 11.84EE192 pKa = 4.15CCIFKK197 pKa = 10.0TPSVEE202 pKa = 4.32CKK204 pKa = 10.28SKK206 pKa = 11.07AVLAGGKK213 pKa = 7.1ITNYY217 pKa = 9.71INTFHH222 pKa = 6.36EE223 pKa = 4.75TIIQCPCAQKK233 pKa = 10.7CVIFINSQIDD243 pKa = 3.55VLISDD248 pKa = 5.59GILIKK253 pKa = 10.28STFEE257 pKa = 4.61LEE259 pKa = 3.88NLHH262 pKa = 6.62NIKK265 pKa = 10.26DD266 pKa = 3.64QHH268 pKa = 5.88TKK270 pKa = 10.69SILRR274 pKa = 11.84QQQPEE279 pKa = 4.1TSKK282 pKa = 11.35LEE284 pKa = 3.94RR285 pKa = 11.84QYY287 pKa = 11.83LEE289 pKa = 4.45FNNNCGDD296 pKa = 4.23SITKK300 pKa = 10.12AGCEE304 pKa = 3.84LEE306 pKa = 4.28TIEE309 pKa = 4.8FSSSIYY315 pKa = 9.31MIHH318 pKa = 6.36NAVMPNTIFKK328 pKa = 8.0TFCAVEE334 pKa = 4.09KK335 pKa = 11.02LPINDD340 pKa = 3.37QLEE343 pKa = 4.21PLNSKK348 pKa = 10.2FSVPATVQLGKK359 pKa = 10.12QRR361 pKa = 11.84YY362 pKa = 7.88IAHH365 pKa = 6.95KK366 pKa = 9.06VNPVTFQLDD375 pKa = 3.67YY376 pKa = 11.43TNDD379 pKa = 3.54PDD381 pKa = 5.41FISATTTTLPTIGSCRR397 pKa = 11.84RR398 pKa = 11.84SDD400 pKa = 3.27GYY402 pKa = 11.09AWHH405 pKa = 7.06GNFSIEE411 pKa = 4.08KK412 pKa = 8.38NEE414 pKa = 4.63ARR416 pKa = 11.84IEE418 pKa = 4.32DD419 pKa = 4.19CTATTLDD426 pKa = 3.95WNPVCNITKK435 pKa = 10.42LEE437 pKa = 4.13PLVKK441 pKa = 10.33VYY443 pKa = 10.78AVDD446 pKa = 3.83KK447 pKa = 10.73KK448 pKa = 9.88ITAKK452 pKa = 10.47GVNCDD457 pKa = 4.78DD458 pKa = 3.48ICPYY462 pKa = 11.04AVLCSEE468 pKa = 4.16EE469 pKa = 4.36QKK471 pKa = 10.67RR472 pKa = 11.84SPIYY476 pKa = 9.19QACLAIEE483 pKa = 4.02QDD485 pKa = 2.87ISTLLKK491 pKa = 10.2VEE493 pKa = 4.55RR494 pKa = 11.84PTSVKK499 pKa = 9.53MAALTKK505 pKa = 9.64TEE507 pKa = 4.32PLAGTIDD514 pKa = 3.62VNFLQEE520 pKa = 4.01VAKK523 pKa = 10.85VQAQRR528 pKa = 11.84FDD530 pKa = 3.74LTVEE534 pKa = 3.99TGLDD538 pKa = 3.38HH539 pKa = 6.65TQVLDD544 pKa = 4.11DD545 pKa = 4.35LFKK548 pKa = 10.88LTVALPQDD556 pKa = 3.56STFFPDD562 pKa = 3.61RR563 pKa = 11.84SQAAIDD569 pKa = 4.32LGLFQNAMFNSKK581 pKa = 10.72ADD583 pKa = 3.72MDD585 pKa = 4.31HH586 pKa = 6.34VAAFPWLAGWRR597 pKa = 11.84FGRR600 pKa = 11.84QINGLSYY607 pKa = 8.05TTSNLIRR614 pKa = 11.84SYY616 pKa = 10.77RR617 pKa = 11.84QFSNTVNKK625 pKa = 10.14NFQQVATALSAMADD639 pKa = 3.97QITMNYY645 pKa = 9.49EE646 pKa = 3.65VTTRR650 pKa = 11.84LYY652 pKa = 10.99EE653 pKa = 4.38SIQRR657 pKa = 11.84SYY659 pKa = 10.36QAYY662 pKa = 8.5EE663 pKa = 3.85NNFAKK668 pKa = 9.69IDD670 pKa = 3.65YY671 pKa = 9.74RR672 pKa = 11.84MSEE675 pKa = 4.21IEE677 pKa = 4.97FMTAKK682 pKa = 10.55LSLINQYY689 pKa = 7.41FTKK692 pKa = 10.34IATRR696 pKa = 11.84QSQLASEE703 pKa = 4.39HH704 pKa = 6.18QIFDD708 pKa = 4.03LRR710 pKa = 11.84VGSCRR715 pKa = 11.84TANPACFGGQGVYY728 pKa = 9.73LAHH731 pKa = 7.42AEE733 pKa = 4.26FEE735 pKa = 4.6TPSQKK740 pKa = 10.94LLIVNYY746 pKa = 10.22LGSTEE751 pKa = 3.85KK752 pKa = 10.65CKK754 pKa = 10.49QVFTANKK761 pKa = 8.74ICTKK765 pKa = 10.5QGTATIAPFGCAFFKK780 pKa = 10.78TSDD783 pKa = 3.47SDD785 pKa = 4.62SEE787 pKa = 4.32VLRR790 pKa = 11.84NLKK793 pKa = 10.67DD794 pKa = 3.28NGPCHH799 pKa = 6.52VPPINVQDD807 pKa = 3.97CQLPTNEE814 pKa = 4.83IIQLQLINYY823 pKa = 9.21QEE825 pKa = 4.11TLEE828 pKa = 4.24LGIRR832 pKa = 11.84QVNLTRR838 pKa = 11.84VEE840 pKa = 4.34FNQKK844 pKa = 8.74IANISTFNNNIKK856 pKa = 10.35QLVNSIDD863 pKa = 3.76KK864 pKa = 10.73VPSLGDD870 pKa = 3.35MLEE873 pKa = 4.17KK874 pKa = 9.8QTIASLNSIIAQINAWSIWDD894 pKa = 3.64IIKK897 pKa = 9.99WFLVCGVIIAFSALGISVIKK917 pKa = 10.68LVTKK921 pKa = 10.58RR922 pKa = 3.56
Molecular weight: 103.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346I7J2|A0A346I7J2_9NIDO Minor membrane protein 2 OS=Bellinger River virus OX=2301728 GN=MM2 PE=4 SV=1
MM1 pKa = 7.34ATFLPMPMQLPLMQPTRR18 pKa = 11.84PVQRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84PAKK30 pKa = 9.55RR31 pKa = 11.84QQRR34 pKa = 11.84NPNNAMTKK42 pKa = 9.96KK43 pKa = 10.6VDD45 pKa = 3.56MLTKK49 pKa = 10.58KK50 pKa = 10.25LDD52 pKa = 3.55KK53 pKa = 10.97LMPTVKK59 pKa = 9.91PDD61 pKa = 3.58PNVYY65 pKa = 9.03TFTAKK70 pKa = 10.51YY71 pKa = 9.98GEE73 pKa = 4.31KK74 pKa = 10.16LAPLLITPEE83 pKa = 3.88QDD85 pKa = 2.87SDD87 pKa = 3.71PRR89 pKa = 11.84NHH91 pKa = 6.39MSQEE95 pKa = 3.77AMTKK99 pKa = 10.01FANDD103 pKa = 2.83VSRR106 pKa = 11.84RR107 pKa = 11.84LKK109 pKa = 10.83AGAGTVTIAPTGVVTVHH126 pKa = 6.34LQFKK130 pKa = 10.04PIALNPNAQAFVPRR144 pKa = 11.84DD145 pKa = 3.35LSEE148 pKa = 3.95VSEE151 pKa = 4.31AA152 pKa = 4.14
MM1 pKa = 7.34ATFLPMPMQLPLMQPTRR18 pKa = 11.84PVQRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84PAKK30 pKa = 9.55RR31 pKa = 11.84QQRR34 pKa = 11.84NPNNAMTKK42 pKa = 9.96KK43 pKa = 10.6VDD45 pKa = 3.56MLTKK49 pKa = 10.58KK50 pKa = 10.25LDD52 pKa = 3.55KK53 pKa = 10.97LMPTVKK59 pKa = 9.91PDD61 pKa = 3.58PNVYY65 pKa = 9.03TFTAKK70 pKa = 10.51YY71 pKa = 9.98GEE73 pKa = 4.31KK74 pKa = 10.16LAPLLITPEE83 pKa = 3.88QDD85 pKa = 2.87SDD87 pKa = 3.71PRR89 pKa = 11.84NHH91 pKa = 6.39MSQEE95 pKa = 3.77AMTKK99 pKa = 10.01FANDD103 pKa = 2.83VSRR106 pKa = 11.84RR107 pKa = 11.84LKK109 pKa = 10.83AGAGTVTIAPTGVVTVHH126 pKa = 6.34LQFKK130 pKa = 10.04PIALNPNAQAFVPRR144 pKa = 11.84DD145 pKa = 3.35LSEE148 pKa = 3.95VSEE151 pKa = 4.31AA152 pKa = 4.14
Molecular weight: 17.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14867 |
152 |
7459 |
1858.4 |
209.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.625 ± 0.181 | 2.556 ± 0.161 |
4.856 ± 0.317 | 4.581 ± 0.214 |
4.365 ± 0.194 | 3.276 ± 0.146 |
3.504 ± 0.41 | 7.305 ± 0.256 |
5.892 ± 0.214 | 8.051 ± 0.135 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.291 ± 0.104 | 7.533 ± 0.196 |
3.787 ± 0.223 | 4.903 ± 0.29 |
3.538 ± 0.103 | 7.984 ± 0.328 |
10.244 ± 0.406 | 5.139 ± 0.312 |
0.888 ± 0.1 | 3.679 ± 0.16 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
