
Kitasatospora setae (strain ATCC 33774 / DSM 43861 / JCM 3304 / KCC A-0304 / NBRC 14216 / KM-6054) (Streptomyces setae)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Kitasatospora; Kitasatospora setae
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
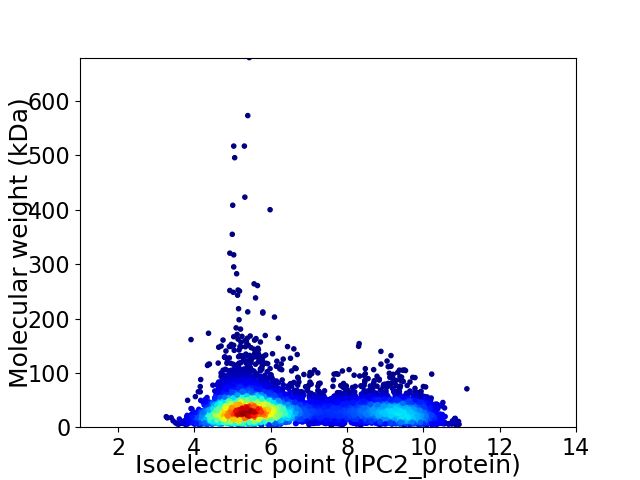
Virtual 2D-PAGE plot for 7443 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E4N2T3|E4N2T3_KITSK Robl_LC7 domain-containing protein OS=Kitasatospora setae (strain ATCC 33774 / DSM 43861 / JCM 3304 / KCC A-0304 / NBRC 14216 / KM-6054) OX=452652 GN=KSE_67090 PE=4 SV=1
MM1 pKa = 7.74AGTGEE6 pKa = 4.21ALEE9 pKa = 4.12VWIDD13 pKa = 3.41QDD15 pKa = 3.99LCTGDD20 pKa = 5.01GICAQYY26 pKa = 10.9APEE29 pKa = 4.23VFEE32 pKa = 5.9LDD34 pKa = 3.05IDD36 pKa = 3.65GLAYY40 pKa = 10.69VKK42 pKa = 11.17GEE44 pKa = 4.07DD45 pKa = 4.75DD46 pKa = 4.81EE47 pKa = 6.03LRR49 pKa = 11.84QQPGQSVPVPLTLLQDD65 pKa = 3.97VVDD68 pKa = 4.33SAKK71 pKa = 10.65DD72 pKa = 3.57CPGDD76 pKa = 4.13CIHH79 pKa = 6.47VRR81 pKa = 11.84RR82 pKa = 11.84AADD85 pKa = 3.45GVEE88 pKa = 4.48VYY90 pKa = 10.92GPDD93 pKa = 3.7AEE95 pKa = 4.31
MM1 pKa = 7.74AGTGEE6 pKa = 4.21ALEE9 pKa = 4.12VWIDD13 pKa = 3.41QDD15 pKa = 3.99LCTGDD20 pKa = 5.01GICAQYY26 pKa = 10.9APEE29 pKa = 4.23VFEE32 pKa = 5.9LDD34 pKa = 3.05IDD36 pKa = 3.65GLAYY40 pKa = 10.69VKK42 pKa = 11.17GEE44 pKa = 4.07DD45 pKa = 4.75DD46 pKa = 4.81EE47 pKa = 6.03LRR49 pKa = 11.84QQPGQSVPVPLTLLQDD65 pKa = 3.97VVDD68 pKa = 4.33SAKK71 pKa = 10.65DD72 pKa = 3.57CPGDD76 pKa = 4.13CIHH79 pKa = 6.47VRR81 pKa = 11.84RR82 pKa = 11.84AADD85 pKa = 3.45GVEE88 pKa = 4.48VYY90 pKa = 10.92GPDD93 pKa = 3.7AEE95 pKa = 4.31
Molecular weight: 10.16 kDa
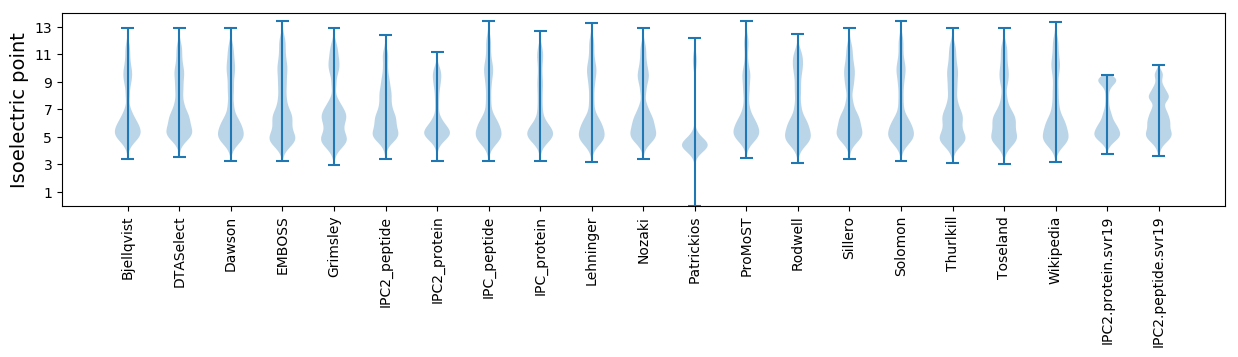
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E4NAR0|E4NAR0_KITSK Uncharacterized protein OS=Kitasatospora setae (strain ATCC 33774 / DSM 43861 / JCM 3304 / KCC A-0304 / NBRC 14216 / KM-6054) OX=452652 GN=KSE_24770 PE=4 SV=1
MM1 pKa = 7.17PQALRR6 pKa = 11.84VRR8 pKa = 11.84RR9 pKa = 11.84VLKK12 pKa = 10.51ARR14 pKa = 11.84QVPKK18 pKa = 10.38AQQALPVLRR27 pKa = 11.84VRR29 pKa = 11.84LATQVLRR36 pKa = 11.84ARR38 pKa = 11.84QALMAPPAPKK48 pKa = 10.09ARR50 pKa = 11.84RR51 pKa = 11.84ARR53 pKa = 11.84QALRR57 pKa = 11.84ARR59 pKa = 11.84RR60 pKa = 11.84VLRR63 pKa = 11.84ARR65 pKa = 11.84PALMVPPVPKK75 pKa = 10.38ARR77 pKa = 11.84QGPLARR83 pKa = 11.84RR84 pKa = 11.84ARR86 pKa = 11.84QALRR90 pKa = 11.84VRR92 pKa = 11.84SARR95 pKa = 11.84QALLVPKK102 pKa = 10.33ARR104 pKa = 11.84RR105 pKa = 11.84ALQAQPVLMAPPVPKK120 pKa = 10.34ARR122 pKa = 11.84QVRR125 pKa = 11.84LATQVPLAPKK135 pKa = 9.98ARR137 pKa = 11.84QALRR141 pKa = 11.84ARR143 pKa = 11.84PAPSALPAPKK153 pKa = 10.05AQRR156 pKa = 11.84ALMARR161 pKa = 11.84QVRR164 pKa = 11.84RR165 pKa = 11.84VRR167 pKa = 11.84PVLRR171 pKa = 11.84VRR173 pKa = 11.84PVLLVLKK180 pKa = 10.1DD181 pKa = 3.87RR182 pKa = 11.84PAILALPVPKK192 pKa = 10.23ARR194 pKa = 11.84RR195 pKa = 11.84VFKK198 pKa = 10.65ALRR201 pKa = 11.84ARR203 pKa = 11.84RR204 pKa = 11.84APKK207 pKa = 9.98DD208 pKa = 3.19QRR210 pKa = 11.84ALMARR215 pKa = 11.84QVRR218 pKa = 11.84RR219 pKa = 11.84ALPALKK225 pKa = 10.2DD226 pKa = 3.9RR227 pKa = 11.84PATPALPAPKK237 pKa = 9.93ARR239 pKa = 11.84RR240 pKa = 11.84ALQARR245 pKa = 11.84QARR248 pKa = 11.84QVRR251 pKa = 11.84RR252 pKa = 11.84VRR254 pKa = 11.84PATRR258 pKa = 11.84VPPAPKK264 pKa = 9.92ARR266 pKa = 11.84QVRR269 pKa = 11.84LATPAPKK276 pKa = 10.05DD277 pKa = 3.42QRR279 pKa = 11.84ALQAQSARR287 pKa = 11.84RR288 pKa = 11.84APRR291 pKa = 11.84ARR293 pKa = 11.84PAPSALPAPKK303 pKa = 10.0ARR305 pKa = 11.84RR306 pKa = 11.84ALQAQPGRR314 pKa = 11.84QVRR317 pKa = 11.84RR318 pKa = 11.84VRR320 pKa = 11.84PATRR324 pKa = 11.84VPPAPKK330 pKa = 10.0ARR332 pKa = 11.84QVQLATPAPKK342 pKa = 10.12DD343 pKa = 3.27QRR345 pKa = 11.84EE346 pKa = 4.06PLARR350 pKa = 11.84QARR353 pKa = 11.84QVRR356 pKa = 11.84RR357 pKa = 11.84VRR359 pKa = 11.84PATRR363 pKa = 11.84VPPAPKK369 pKa = 9.92ARR371 pKa = 11.84QVRR374 pKa = 11.84LATPAPKK381 pKa = 10.14DD382 pKa = 3.47RR383 pKa = 11.84PARR386 pKa = 11.84RR387 pKa = 11.84APRR390 pKa = 11.84ARR392 pKa = 11.84RR393 pKa = 11.84ALRR396 pKa = 11.84ARR398 pKa = 11.84PARR401 pKa = 11.84QVRR404 pKa = 11.84PATRR408 pKa = 11.84VPPAPKK414 pKa = 9.92ARR416 pKa = 11.84QVRR419 pKa = 11.84LATPAPKK426 pKa = 10.09DD427 pKa = 3.27QRR429 pKa = 11.84EE430 pKa = 4.06PLARR434 pKa = 11.84QARR437 pKa = 11.84QVRR440 pKa = 11.84QAPKK444 pKa = 9.57AQQALRR450 pKa = 11.84ARR452 pKa = 11.84PARR455 pKa = 11.84RR456 pKa = 11.84VLPALRR462 pKa = 11.84GRR464 pKa = 11.84PATQVRR470 pKa = 11.84RR471 pKa = 11.84VLRR474 pKa = 11.84ARR476 pKa = 11.84SALPVLKK483 pKa = 10.44DD484 pKa = 3.91RR485 pKa = 11.84PATPALPAPKK495 pKa = 9.75DD496 pKa = 3.63RR497 pKa = 11.84RR498 pKa = 11.84VLRR501 pKa = 11.84AQQARR506 pKa = 11.84RR507 pKa = 11.84APRR510 pKa = 11.84AQPAPSALPALPAPKK525 pKa = 9.81DD526 pKa = 3.49RR527 pKa = 11.84PARR530 pKa = 11.84RR531 pKa = 11.84VLPALRR537 pKa = 11.84GRR539 pKa = 11.84PATQVRR545 pKa = 11.84RR546 pKa = 11.84VLRR549 pKa = 11.84ARR551 pKa = 11.84SALPVLKK558 pKa = 10.44DD559 pKa = 3.91RR560 pKa = 11.84PATPALPAPKK570 pKa = 9.84DD571 pKa = 3.78RR572 pKa = 11.84QVLRR576 pKa = 11.84ARR578 pKa = 11.84QARR581 pKa = 11.84RR582 pKa = 11.84APRR585 pKa = 11.84GRR587 pKa = 11.84PAPSVLLARR596 pKa = 11.84RR597 pKa = 11.84VPAARR602 pKa = 11.84PVLLARR608 pKa = 11.84RR609 pKa = 11.84VRR611 pKa = 11.84PGRR614 pKa = 11.84RR615 pKa = 11.84VPLAPPVPGGRR626 pKa = 11.84PGRR629 pKa = 11.84PGRR632 pKa = 11.84PGCRR636 pKa = 11.84II637 pKa = 3.33
MM1 pKa = 7.17PQALRR6 pKa = 11.84VRR8 pKa = 11.84RR9 pKa = 11.84VLKK12 pKa = 10.51ARR14 pKa = 11.84QVPKK18 pKa = 10.38AQQALPVLRR27 pKa = 11.84VRR29 pKa = 11.84LATQVLRR36 pKa = 11.84ARR38 pKa = 11.84QALMAPPAPKK48 pKa = 10.09ARR50 pKa = 11.84RR51 pKa = 11.84ARR53 pKa = 11.84QALRR57 pKa = 11.84ARR59 pKa = 11.84RR60 pKa = 11.84VLRR63 pKa = 11.84ARR65 pKa = 11.84PALMVPPVPKK75 pKa = 10.38ARR77 pKa = 11.84QGPLARR83 pKa = 11.84RR84 pKa = 11.84ARR86 pKa = 11.84QALRR90 pKa = 11.84VRR92 pKa = 11.84SARR95 pKa = 11.84QALLVPKK102 pKa = 10.33ARR104 pKa = 11.84RR105 pKa = 11.84ALQAQPVLMAPPVPKK120 pKa = 10.34ARR122 pKa = 11.84QVRR125 pKa = 11.84LATQVPLAPKK135 pKa = 9.98ARR137 pKa = 11.84QALRR141 pKa = 11.84ARR143 pKa = 11.84PAPSALPAPKK153 pKa = 10.05AQRR156 pKa = 11.84ALMARR161 pKa = 11.84QVRR164 pKa = 11.84RR165 pKa = 11.84VRR167 pKa = 11.84PVLRR171 pKa = 11.84VRR173 pKa = 11.84PVLLVLKK180 pKa = 10.1DD181 pKa = 3.87RR182 pKa = 11.84PAILALPVPKK192 pKa = 10.23ARR194 pKa = 11.84RR195 pKa = 11.84VFKK198 pKa = 10.65ALRR201 pKa = 11.84ARR203 pKa = 11.84RR204 pKa = 11.84APKK207 pKa = 9.98DD208 pKa = 3.19QRR210 pKa = 11.84ALMARR215 pKa = 11.84QVRR218 pKa = 11.84RR219 pKa = 11.84ALPALKK225 pKa = 10.2DD226 pKa = 3.9RR227 pKa = 11.84PATPALPAPKK237 pKa = 9.93ARR239 pKa = 11.84RR240 pKa = 11.84ALQARR245 pKa = 11.84QARR248 pKa = 11.84QVRR251 pKa = 11.84RR252 pKa = 11.84VRR254 pKa = 11.84PATRR258 pKa = 11.84VPPAPKK264 pKa = 9.92ARR266 pKa = 11.84QVRR269 pKa = 11.84LATPAPKK276 pKa = 10.05DD277 pKa = 3.42QRR279 pKa = 11.84ALQAQSARR287 pKa = 11.84RR288 pKa = 11.84APRR291 pKa = 11.84ARR293 pKa = 11.84PAPSALPAPKK303 pKa = 10.0ARR305 pKa = 11.84RR306 pKa = 11.84ALQAQPGRR314 pKa = 11.84QVRR317 pKa = 11.84RR318 pKa = 11.84VRR320 pKa = 11.84PATRR324 pKa = 11.84VPPAPKK330 pKa = 10.0ARR332 pKa = 11.84QVQLATPAPKK342 pKa = 10.12DD343 pKa = 3.27QRR345 pKa = 11.84EE346 pKa = 4.06PLARR350 pKa = 11.84QARR353 pKa = 11.84QVRR356 pKa = 11.84RR357 pKa = 11.84VRR359 pKa = 11.84PATRR363 pKa = 11.84VPPAPKK369 pKa = 9.92ARR371 pKa = 11.84QVRR374 pKa = 11.84LATPAPKK381 pKa = 10.14DD382 pKa = 3.47RR383 pKa = 11.84PARR386 pKa = 11.84RR387 pKa = 11.84APRR390 pKa = 11.84ARR392 pKa = 11.84RR393 pKa = 11.84ALRR396 pKa = 11.84ARR398 pKa = 11.84PARR401 pKa = 11.84QVRR404 pKa = 11.84PATRR408 pKa = 11.84VPPAPKK414 pKa = 9.92ARR416 pKa = 11.84QVRR419 pKa = 11.84LATPAPKK426 pKa = 10.09DD427 pKa = 3.27QRR429 pKa = 11.84EE430 pKa = 4.06PLARR434 pKa = 11.84QARR437 pKa = 11.84QVRR440 pKa = 11.84QAPKK444 pKa = 9.57AQQALRR450 pKa = 11.84ARR452 pKa = 11.84PARR455 pKa = 11.84RR456 pKa = 11.84VLPALRR462 pKa = 11.84GRR464 pKa = 11.84PATQVRR470 pKa = 11.84RR471 pKa = 11.84VLRR474 pKa = 11.84ARR476 pKa = 11.84SALPVLKK483 pKa = 10.44DD484 pKa = 3.91RR485 pKa = 11.84PATPALPAPKK495 pKa = 9.75DD496 pKa = 3.63RR497 pKa = 11.84RR498 pKa = 11.84VLRR501 pKa = 11.84AQQARR506 pKa = 11.84RR507 pKa = 11.84APRR510 pKa = 11.84AQPAPSALPALPAPKK525 pKa = 9.81DD526 pKa = 3.49RR527 pKa = 11.84PARR530 pKa = 11.84RR531 pKa = 11.84VLPALRR537 pKa = 11.84GRR539 pKa = 11.84PATQVRR545 pKa = 11.84RR546 pKa = 11.84VLRR549 pKa = 11.84ARR551 pKa = 11.84SALPVLKK558 pKa = 10.44DD559 pKa = 3.91RR560 pKa = 11.84PATPALPAPKK570 pKa = 9.84DD571 pKa = 3.78RR572 pKa = 11.84QVLRR576 pKa = 11.84ARR578 pKa = 11.84QARR581 pKa = 11.84RR582 pKa = 11.84APRR585 pKa = 11.84GRR587 pKa = 11.84PAPSVLLARR596 pKa = 11.84RR597 pKa = 11.84VPAARR602 pKa = 11.84PVLLARR608 pKa = 11.84RR609 pKa = 11.84VRR611 pKa = 11.84PGRR614 pKa = 11.84RR615 pKa = 11.84VPLAPPVPGGRR626 pKa = 11.84PGRR629 pKa = 11.84PGRR632 pKa = 11.84PGCRR636 pKa = 11.84II637 pKa = 3.33
Molecular weight: 70.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2504147 |
29 |
6512 |
336.4 |
35.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.001 ± 0.054 | 0.762 ± 0.007 |
5.785 ± 0.022 | 5.324 ± 0.033 |
2.544 ± 0.014 | 9.748 ± 0.031 |
2.298 ± 0.015 | 2.59 ± 0.02 |
1.705 ± 0.023 | 11.03 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.435 ± 0.013 | 1.662 ± 0.019 |
6.767 ± 0.031 | 2.716 ± 0.018 |
8.279 ± 0.036 | 4.692 ± 0.024 |
6.022 ± 0.038 | 8.124 ± 0.025 |
1.528 ± 0.012 | 1.985 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
