
Seal anellovirus 2
Taxonomy: Viruses; Anelloviridae; Lambdatorquevirus; Torque teno pinniped virus 2
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
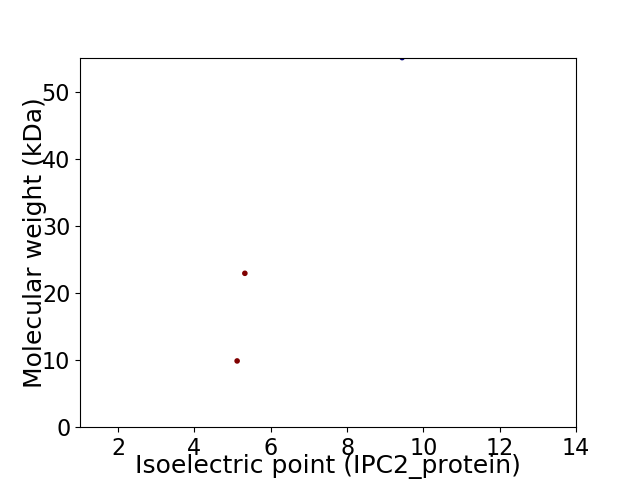
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
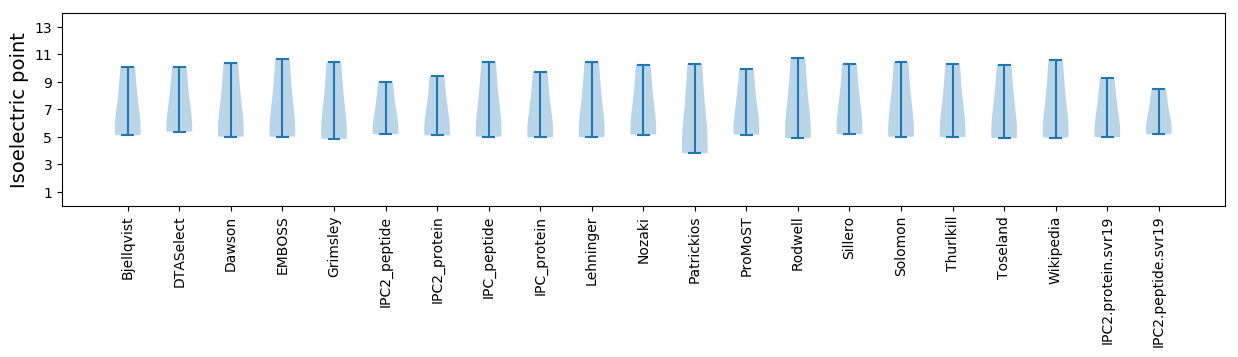
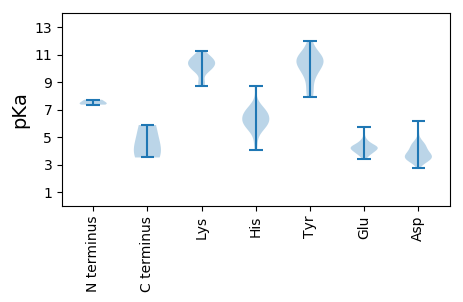
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V5NF43|V5NF43_9VIRU ORF2 OS=Seal anellovirus 2 OX=1427157 PE=4 SV=1
MM1 pKa = 7.71SADD4 pKa = 3.85CDD6 pKa = 3.96SARR9 pKa = 11.84GPDD12 pKa = 3.44LCHH15 pKa = 6.06PQLYY19 pKa = 9.75KK20 pKa = 10.99YY21 pKa = 10.59KK22 pKa = 9.05EE23 pKa = 4.38AKK25 pKa = 8.85WKK27 pKa = 9.11QQVSKK32 pKa = 10.24LHH34 pKa = 7.35RR35 pKa = 11.84EE36 pKa = 3.83FCGCNNFLDD45 pKa = 4.05HH46 pKa = 6.71FKK48 pKa = 10.61WPGTALPGGGDD59 pKa = 3.48AGEE62 pKa = 4.65GSQDD66 pKa = 3.41GGDD69 pKa = 3.36TAGIGGLSTTGGGGGDD85 pKa = 4.54SITDD89 pKa = 3.57MEE91 pKa = 4.71LLRR94 pKa = 5.87
MM1 pKa = 7.71SADD4 pKa = 3.85CDD6 pKa = 3.96SARR9 pKa = 11.84GPDD12 pKa = 3.44LCHH15 pKa = 6.06PQLYY19 pKa = 9.75KK20 pKa = 10.99YY21 pKa = 10.59KK22 pKa = 9.05EE23 pKa = 4.38AKK25 pKa = 8.85WKK27 pKa = 9.11QQVSKK32 pKa = 10.24LHH34 pKa = 7.35RR35 pKa = 11.84EE36 pKa = 3.83FCGCNNFLDD45 pKa = 4.05HH46 pKa = 6.71FKK48 pKa = 10.61WPGTALPGGGDD59 pKa = 3.48AGEE62 pKa = 4.65GSQDD66 pKa = 3.41GGDD69 pKa = 3.36TAGIGGLSTTGGGGGDD85 pKa = 4.54SITDD89 pKa = 3.57MEE91 pKa = 4.71LLRR94 pKa = 5.87
Molecular weight: 9.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V5NF43|V5NF43_9VIRU ORF2 OS=Seal anellovirus 2 OX=1427157 PE=4 SV=1
MM1 pKa = 7.36ARR3 pKa = 11.84HH4 pKa = 6.13RR5 pKa = 11.84FTWRR9 pKa = 11.84GRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FPRR18 pKa = 11.84WRR20 pKa = 11.84RR21 pKa = 11.84YY22 pKa = 8.31RR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 8.97WRR27 pKa = 11.84PFHH30 pKa = 5.69HH31 pKa = 6.18WRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84FHH39 pKa = 6.66HH40 pKa = 5.7RR41 pKa = 11.84HH42 pKa = 4.09GTASVRR48 pKa = 11.84YY49 pKa = 7.88WRR51 pKa = 11.84SSRR54 pKa = 11.84NKK56 pKa = 8.73TIVVRR61 pKa = 11.84GWEE64 pKa = 4.1PLGNICPGSDD74 pKa = 3.07ASKK77 pKa = 10.53EE78 pKa = 3.98ATPWEE83 pKa = 4.35VLDD86 pKa = 5.4KK87 pKa = 10.92DD88 pKa = 4.31YY89 pKa = 11.61EE90 pKa = 4.22DD91 pKa = 3.92TKK93 pKa = 11.09TKK95 pKa = 10.64GKK97 pKa = 10.55EE98 pKa = 3.66SDD100 pKa = 3.83CAKK103 pKa = 10.18SCWSGTFGGHH113 pKa = 6.55FFTFKK118 pKa = 11.06LLLQRR123 pKa = 11.84AKK125 pKa = 10.35IRR127 pKa = 11.84LCQFSSDD134 pKa = 3.52WNGWDD139 pKa = 3.28YY140 pKa = 11.98LEE142 pKa = 5.08FKK144 pKa = 10.45GGYY147 pKa = 8.22IWLPRR152 pKa = 11.84LAGLQWMFNKK162 pKa = 10.17DD163 pKa = 3.42HH164 pKa = 6.85SIKK167 pKa = 10.96NPTEE171 pKa = 3.95TKK173 pKa = 10.24EE174 pKa = 4.21GQEE177 pKa = 3.69LAAAKK182 pKa = 9.91SWTHH186 pKa = 5.69PGVWLNRR193 pKa = 11.84RR194 pKa = 11.84GPIIMHH200 pKa = 7.04DD201 pKa = 3.6PNLFPSHH208 pKa = 6.71RR209 pKa = 11.84GFRR212 pKa = 11.84RR213 pKa = 11.84LKK215 pKa = 10.03VKK217 pKa = 10.39PPTGWEE223 pKa = 3.84GHH225 pKa = 5.63YY226 pKa = 10.71SLPAVMNYY234 pKa = 9.38IFFQWFWTALDD245 pKa = 4.09MQHH248 pKa = 7.02AMYY251 pKa = 10.25DD252 pKa = 3.65YY253 pKa = 10.16WCVKK257 pKa = 9.83QNPTAEE263 pKa = 4.2HH264 pKa = 6.12TKK266 pKa = 9.75ICCQVPWWAAQIDD279 pKa = 4.39DD280 pKa = 3.75TWRR283 pKa = 11.84PKK285 pKa = 10.76YY286 pKa = 10.86ADD288 pKa = 4.73DD289 pKa = 6.16DD290 pKa = 4.21PAKK293 pKa = 9.98TCGGDD298 pKa = 3.96VRR300 pKa = 11.84EE301 pKa = 4.25LWTDD305 pKa = 3.59RR306 pKa = 11.84EE307 pKa = 4.33NYY309 pKa = 9.63LQSKK313 pKa = 10.29CNTNQARR320 pKa = 11.84TDD322 pKa = 3.6QNNIPANTQNWGPFLPQSFVRR343 pKa = 11.84PGEE346 pKa = 4.12AGFSLWFKK354 pKa = 10.55YY355 pKa = 10.22KK356 pKa = 10.46FYY358 pKa = 10.65FQLSGDD364 pKa = 3.61SVYY367 pKa = 10.61RR368 pKa = 11.84QQPHH372 pKa = 6.26TDD374 pKa = 3.22PKK376 pKa = 10.82VVIPRR381 pKa = 11.84APKK384 pKa = 10.38AKK386 pKa = 8.81NTLTGEE392 pKa = 4.25VQPGPEE398 pKa = 4.11RR399 pKa = 11.84PQDD402 pKa = 3.3PWDD405 pKa = 4.12ILPGDD410 pKa = 4.5LDD412 pKa = 3.29RR413 pKa = 11.84HH414 pKa = 6.26GILKK418 pKa = 9.57EE419 pKa = 3.41RR420 pKa = 11.84AYY422 pKa = 10.9RR423 pKa = 11.84RR424 pKa = 11.84ITSTGPGDD432 pKa = 3.75EE433 pKa = 4.14PAKK436 pKa = 10.24VGPSRR441 pKa = 11.84VRR443 pKa = 11.84FRR445 pKa = 11.84SDD447 pKa = 2.72PSVRR451 pKa = 11.84RR452 pKa = 11.84KK453 pKa = 9.64RR454 pKa = 11.84LRR456 pKa = 11.84HH457 pKa = 4.86ILSRR461 pKa = 11.84LLEE464 pKa = 4.2
MM1 pKa = 7.36ARR3 pKa = 11.84HH4 pKa = 6.13RR5 pKa = 11.84FTWRR9 pKa = 11.84GRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FPRR18 pKa = 11.84WRR20 pKa = 11.84RR21 pKa = 11.84YY22 pKa = 8.31RR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 8.97WRR27 pKa = 11.84PFHH30 pKa = 5.69HH31 pKa = 6.18WRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84FHH39 pKa = 6.66HH40 pKa = 5.7RR41 pKa = 11.84HH42 pKa = 4.09GTASVRR48 pKa = 11.84YY49 pKa = 7.88WRR51 pKa = 11.84SSRR54 pKa = 11.84NKK56 pKa = 8.73TIVVRR61 pKa = 11.84GWEE64 pKa = 4.1PLGNICPGSDD74 pKa = 3.07ASKK77 pKa = 10.53EE78 pKa = 3.98ATPWEE83 pKa = 4.35VLDD86 pKa = 5.4KK87 pKa = 10.92DD88 pKa = 4.31YY89 pKa = 11.61EE90 pKa = 4.22DD91 pKa = 3.92TKK93 pKa = 11.09TKK95 pKa = 10.64GKK97 pKa = 10.55EE98 pKa = 3.66SDD100 pKa = 3.83CAKK103 pKa = 10.18SCWSGTFGGHH113 pKa = 6.55FFTFKK118 pKa = 11.06LLLQRR123 pKa = 11.84AKK125 pKa = 10.35IRR127 pKa = 11.84LCQFSSDD134 pKa = 3.52WNGWDD139 pKa = 3.28YY140 pKa = 11.98LEE142 pKa = 5.08FKK144 pKa = 10.45GGYY147 pKa = 8.22IWLPRR152 pKa = 11.84LAGLQWMFNKK162 pKa = 10.17DD163 pKa = 3.42HH164 pKa = 6.85SIKK167 pKa = 10.96NPTEE171 pKa = 3.95TKK173 pKa = 10.24EE174 pKa = 4.21GQEE177 pKa = 3.69LAAAKK182 pKa = 9.91SWTHH186 pKa = 5.69PGVWLNRR193 pKa = 11.84RR194 pKa = 11.84GPIIMHH200 pKa = 7.04DD201 pKa = 3.6PNLFPSHH208 pKa = 6.71RR209 pKa = 11.84GFRR212 pKa = 11.84RR213 pKa = 11.84LKK215 pKa = 10.03VKK217 pKa = 10.39PPTGWEE223 pKa = 3.84GHH225 pKa = 5.63YY226 pKa = 10.71SLPAVMNYY234 pKa = 9.38IFFQWFWTALDD245 pKa = 4.09MQHH248 pKa = 7.02AMYY251 pKa = 10.25DD252 pKa = 3.65YY253 pKa = 10.16WCVKK257 pKa = 9.83QNPTAEE263 pKa = 4.2HH264 pKa = 6.12TKK266 pKa = 9.75ICCQVPWWAAQIDD279 pKa = 4.39DD280 pKa = 3.75TWRR283 pKa = 11.84PKK285 pKa = 10.76YY286 pKa = 10.86ADD288 pKa = 4.73DD289 pKa = 6.16DD290 pKa = 4.21PAKK293 pKa = 9.98TCGGDD298 pKa = 3.96VRR300 pKa = 11.84EE301 pKa = 4.25LWTDD305 pKa = 3.59RR306 pKa = 11.84EE307 pKa = 4.33NYY309 pKa = 9.63LQSKK313 pKa = 10.29CNTNQARR320 pKa = 11.84TDD322 pKa = 3.6QNNIPANTQNWGPFLPQSFVRR343 pKa = 11.84PGEE346 pKa = 4.12AGFSLWFKK354 pKa = 10.55YY355 pKa = 10.22KK356 pKa = 10.46FYY358 pKa = 10.65FQLSGDD364 pKa = 3.61SVYY367 pKa = 10.61RR368 pKa = 11.84QQPHH372 pKa = 6.26TDD374 pKa = 3.22PKK376 pKa = 10.82VVIPRR381 pKa = 11.84APKK384 pKa = 10.38AKK386 pKa = 8.81NTLTGEE392 pKa = 4.25VQPGPEE398 pKa = 4.11RR399 pKa = 11.84PQDD402 pKa = 3.3PWDD405 pKa = 4.12ILPGDD410 pKa = 4.5LDD412 pKa = 3.29RR413 pKa = 11.84HH414 pKa = 6.26GILKK418 pKa = 9.57EE419 pKa = 3.41RR420 pKa = 11.84AYY422 pKa = 10.9RR423 pKa = 11.84RR424 pKa = 11.84ITSTGPGDD432 pKa = 3.75EE433 pKa = 4.14PAKK436 pKa = 10.24VGPSRR441 pKa = 11.84VRR443 pKa = 11.84FRR445 pKa = 11.84SDD447 pKa = 2.72PSVRR451 pKa = 11.84RR452 pKa = 11.84KK453 pKa = 9.64RR454 pKa = 11.84LRR456 pKa = 11.84HH457 pKa = 4.86ILSRR461 pKa = 11.84LLEE464 pKa = 4.2
Molecular weight: 55.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
762 |
94 |
464 |
254.0 |
29.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.856 ± 0.775 | 1.969 ± 0.572 |
6.168 ± 0.788 | 5.381 ± 1.414 |
3.675 ± 1.219 | 8.005 ± 2.578 |
3.018 ± 0.657 | 3.806 ± 0.659 |
6.168 ± 0.115 | 8.005 ± 1.682 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.706 ± 0.375 | 3.543 ± 0.329 |
6.955 ± 0.688 | 4.331 ± 0.242 |
8.793 ± 2.202 | 7.087 ± 1.972 |
6.43 ± 0.97 | 3.15 ± 0.692 |
3.937 ± 1.306 | 3.018 ± 0.388 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
