
Methanomassiliicoccaceae archaeon DOK
Taxonomy: cellular organisms; Archaea; Candidatus Thermoplasmatota; Thermoplasmata; Methanomassiliicoccales; Methanomassiliicoccaceae; unclassified Methanomassiliicoccaceae
Average proteome isoelectric point is 5.49
Get precalculated fractions of proteins
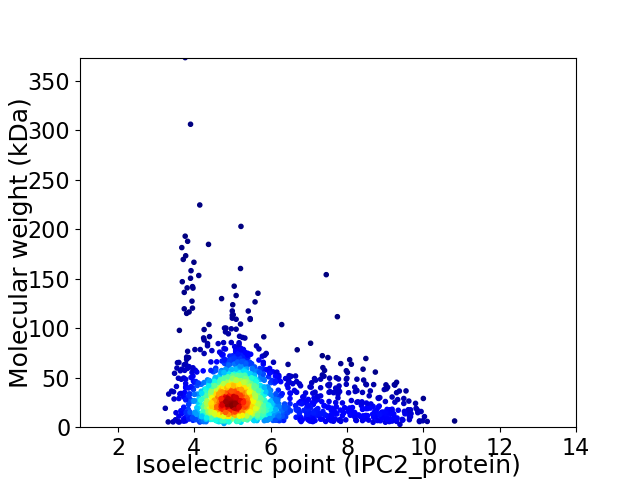
Virtual 2D-PAGE plot for 1835 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
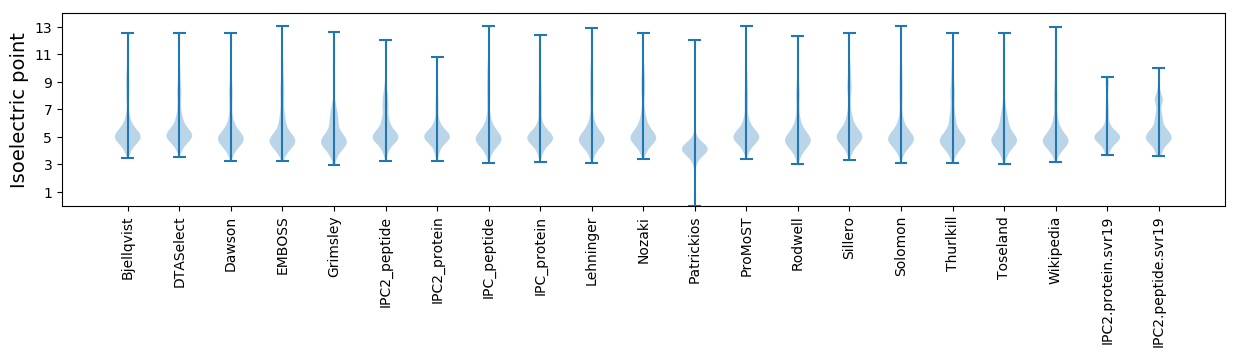
Protein with the lowest isoelectric point:
>tr|A0A6B9TCW7|A0A6B9TCW7_9ARCH Flavodoxin OS=Methanomassiliicoccaceae archaeon DOK OX=1535962 GN=JS82_08600 PE=4 SV=1
MM1 pKa = 7.56NSITKK6 pKa = 10.36AIALLVVLLTATAGVAMMADD26 pKa = 3.59SSDD29 pKa = 3.56AAPADD34 pKa = 3.32IQEE37 pKa = 4.83PVPASNPEE45 pKa = 3.91AGTAIAKK52 pKa = 9.99VDD54 pKa = 3.54GTPYY58 pKa = 10.5TDD60 pKa = 4.33FEE62 pKa = 4.83VAVDD66 pKa = 3.91AAISSNKK73 pKa = 8.73TLTLLNDD80 pKa = 3.66YY81 pKa = 10.88QITNLNLDD89 pKa = 4.16AEE91 pKa = 4.74SVTIDD96 pKa = 4.35LNTKK100 pKa = 8.82ILSFEE105 pKa = 4.31RR106 pKa = 11.84IKK108 pKa = 10.96LINASSLTFEE118 pKa = 4.56NGTLNGLYY126 pKa = 9.97IGAANEE132 pKa = 3.54ASIAVNTGCSVTLNDD147 pKa = 3.04VNYY150 pKa = 7.86YY151 pKa = 9.56TKK153 pKa = 10.35ATGLFPSGDD162 pKa = 3.61AASVTVKK169 pKa = 10.65NGSVVHH175 pKa = 6.61AVGFCIGTNAGSSSNYY191 pKa = 9.64YY192 pKa = 10.43VDD194 pKa = 4.63IKK196 pKa = 10.88ISDD199 pKa = 3.52SGIYY203 pKa = 10.13ADD205 pKa = 4.78SDD207 pKa = 3.64SSAAGTAVFYY217 pKa = 10.33NVPGSLTITDD227 pKa = 3.95STVQGYY233 pKa = 7.01FHH235 pKa = 6.74GVIVRR240 pKa = 11.84GGTAVISGSTITNTMNLSGWEE261 pKa = 4.08TYY263 pKa = 10.59YY264 pKa = 11.09DD265 pKa = 3.83SEE267 pKa = 4.19PWGTGNSIKK276 pKa = 10.7LGALVLGNRR285 pKa = 11.84DD286 pKa = 3.44NASYY290 pKa = 11.03NYY292 pKa = 7.54PTDD295 pKa = 3.19VTLIDD300 pKa = 3.82TDD302 pKa = 4.18VISDD306 pKa = 3.63GTYY309 pKa = 8.67GANYY313 pKa = 8.95SAVYY317 pKa = 8.78MWANPDD323 pKa = 3.56PEE325 pKa = 5.03NGVNLVYY332 pKa = 10.88DD333 pKa = 4.14SACTFSNLNSTTNDD347 pKa = 2.79LFFYY351 pKa = 10.44GNNGANITAGLAVAQAGDD369 pKa = 4.06RR370 pKa = 11.84IYY372 pKa = 11.46NSLQAAIDD380 pKa = 4.12DD381 pKa = 4.35DD382 pKa = 4.48NEE384 pKa = 4.33EE385 pKa = 4.01ITLVGDD391 pKa = 3.42TKK393 pKa = 11.34EE394 pKa = 4.07NVVVDD399 pKa = 4.52GQTVTLDD406 pKa = 3.77LNGKK410 pKa = 6.77TVTNVDD416 pKa = 3.23DD417 pKa = 3.5HH418 pKa = 6.77TITVKK423 pKa = 10.67SGEE426 pKa = 4.21LTVKK430 pKa = 10.76GKK432 pKa = 8.76GTVQNLTHH440 pKa = 6.32QKK442 pKa = 10.27AALAAYY448 pKa = 7.92PGTKK452 pKa = 8.63VTLNGGTFDD461 pKa = 3.78RR462 pKa = 11.84TNEE465 pKa = 4.15TGTGPDD471 pKa = 3.12TSGNNSWYY479 pKa = 9.26TIKK482 pKa = 10.72NHH484 pKa = 6.2GDD486 pKa = 3.24MTIDD490 pKa = 3.39GATVRR495 pKa = 11.84NGGGFSSCINNGWYY509 pKa = 10.05SGSAQGTPGNNDD521 pKa = 3.29LAVYY525 pKa = 10.48AGTNAKK531 pKa = 8.59LTIVSGKK538 pKa = 9.79IDD540 pKa = 3.59GGLNAVKK547 pKa = 10.5NDD549 pKa = 3.86DD550 pKa = 3.85YY551 pKa = 12.05GEE553 pKa = 4.42LLVQGGEE560 pKa = 4.33LTNSVQATILNWNIATIEE578 pKa = 4.16KK579 pKa = 9.02GTFSATSDD587 pKa = 3.24AKK589 pKa = 11.01AVILNGKK596 pKa = 9.18IDD598 pKa = 3.43GTMDD602 pKa = 3.28KK603 pKa = 11.03GQLTISDD610 pKa = 4.06GTFSGPVGIQMMSGGVGIGDD630 pKa = 3.48VTITGGTFNISSDD643 pKa = 4.04NIIDD647 pKa = 4.22PLTGTEE653 pKa = 4.17TNVQVSGGSFSDD665 pKa = 4.12DD666 pKa = 2.96SVLAYY671 pKa = 10.43LVEE674 pKa = 4.46GKK676 pKa = 10.84GLVEE680 pKa = 4.5NGTMFDD686 pKa = 3.52VVDD689 pKa = 3.8VHH691 pKa = 5.86TVTYY695 pKa = 9.09TFNDD699 pKa = 3.14TTYY702 pKa = 9.8IEE704 pKa = 4.53KK705 pKa = 10.73VISGQTATQNTIPALPTLPEE725 pKa = 4.01GCEE728 pKa = 3.79YY729 pKa = 11.26DD730 pKa = 3.44FGEE733 pKa = 4.19WDD735 pKa = 3.08KK736 pKa = 10.37TAPVTEE742 pKa = 4.42DD743 pKa = 2.98VSVSVKK749 pKa = 11.1AEE751 pKa = 3.87ITSLTVAIGTSTVDD765 pKa = 3.34GVTTLTATVDD775 pKa = 3.64STASGTPAYY784 pKa = 8.03TWSSGEE790 pKa = 3.95ATEE793 pKa = 5.49SITPSVGGTYY803 pKa = 10.62DD804 pKa = 3.25VTVTLTVTSGDD815 pKa = 3.44VTVTGKK821 pKa = 10.8ANASVTYY828 pKa = 9.02TPSEE832 pKa = 4.32SGSTTTTTTNPDD844 pKa = 2.85GSTTTVEE851 pKa = 4.1TDD853 pKa = 2.56SDD855 pKa = 4.23GSVTEE860 pKa = 4.45TTTKK864 pKa = 10.63TEE866 pKa = 3.93TTEE869 pKa = 3.96TGNEE873 pKa = 3.66ITTVTTTGSDD883 pKa = 3.36PSGSQTTSSTEE894 pKa = 3.81VTIEE898 pKa = 3.99AASGTSGSVSLPEE911 pKa = 3.92EE912 pKa = 4.36DD913 pKa = 3.6VNSVIEE919 pKa = 4.56SIASAGTTTNTISVNIGEE937 pKa = 4.66NDD939 pKa = 3.65TVTMGSAAISAIAEE953 pKa = 4.24SNAVLEE959 pKa = 4.38ISNEE963 pKa = 3.94NATVVADD970 pKa = 3.66GDD972 pKa = 4.19VIEE975 pKa = 4.64TLSTGDD981 pKa = 3.13VSITVGSATHH991 pKa = 7.22DD992 pKa = 3.74NLNEE996 pKa = 4.1SQQQSVPEE1004 pKa = 3.81NSTIVEE1010 pKa = 4.23LSATVGTSPVHH1021 pKa = 6.39DD1022 pKa = 4.39LNGTVEE1028 pKa = 4.09VTMTYY1033 pKa = 9.25TLPSGIDD1040 pKa = 3.35ADD1042 pKa = 4.2DD1043 pKa = 4.62VIVFYY1048 pKa = 11.37VDD1050 pKa = 4.06DD1051 pKa = 5.87DD1052 pKa = 4.5GALHH1056 pKa = 6.49QMRR1059 pKa = 11.84TVYY1062 pKa = 10.91AEE1064 pKa = 4.86GILTFYY1070 pKa = 7.38TTHH1073 pKa = 6.24FSYY1076 pKa = 11.22YY1077 pKa = 9.83FVGDD1081 pKa = 3.17RR1082 pKa = 11.84SMIPSVDD1089 pKa = 3.84PEE1091 pKa = 4.26PSPGEE1096 pKa = 3.9DD1097 pKa = 3.45DD1098 pKa = 3.78QPVVVPPVDD1107 pKa = 5.25DD1108 pKa = 5.45DD1109 pKa = 4.75DD1110 pKa = 7.13DD1111 pKa = 4.39YY1112 pKa = 12.26VPLPPIVDD1120 pKa = 3.66TDD1122 pKa = 3.89GSGSSGGDD1130 pKa = 3.11DD1131 pKa = 3.19TVKK1134 pKa = 10.49IVACAAAAVVAALMAAFLIISRR1156 pKa = 11.84RR1157 pKa = 11.84DD1158 pKa = 3.09
MM1 pKa = 7.56NSITKK6 pKa = 10.36AIALLVVLLTATAGVAMMADD26 pKa = 3.59SSDD29 pKa = 3.56AAPADD34 pKa = 3.32IQEE37 pKa = 4.83PVPASNPEE45 pKa = 3.91AGTAIAKK52 pKa = 9.99VDD54 pKa = 3.54GTPYY58 pKa = 10.5TDD60 pKa = 4.33FEE62 pKa = 4.83VAVDD66 pKa = 3.91AAISSNKK73 pKa = 8.73TLTLLNDD80 pKa = 3.66YY81 pKa = 10.88QITNLNLDD89 pKa = 4.16AEE91 pKa = 4.74SVTIDD96 pKa = 4.35LNTKK100 pKa = 8.82ILSFEE105 pKa = 4.31RR106 pKa = 11.84IKK108 pKa = 10.96LINASSLTFEE118 pKa = 4.56NGTLNGLYY126 pKa = 9.97IGAANEE132 pKa = 3.54ASIAVNTGCSVTLNDD147 pKa = 3.04VNYY150 pKa = 7.86YY151 pKa = 9.56TKK153 pKa = 10.35ATGLFPSGDD162 pKa = 3.61AASVTVKK169 pKa = 10.65NGSVVHH175 pKa = 6.61AVGFCIGTNAGSSSNYY191 pKa = 9.64YY192 pKa = 10.43VDD194 pKa = 4.63IKK196 pKa = 10.88ISDD199 pKa = 3.52SGIYY203 pKa = 10.13ADD205 pKa = 4.78SDD207 pKa = 3.64SSAAGTAVFYY217 pKa = 10.33NVPGSLTITDD227 pKa = 3.95STVQGYY233 pKa = 7.01FHH235 pKa = 6.74GVIVRR240 pKa = 11.84GGTAVISGSTITNTMNLSGWEE261 pKa = 4.08TYY263 pKa = 10.59YY264 pKa = 11.09DD265 pKa = 3.83SEE267 pKa = 4.19PWGTGNSIKK276 pKa = 10.7LGALVLGNRR285 pKa = 11.84DD286 pKa = 3.44NASYY290 pKa = 11.03NYY292 pKa = 7.54PTDD295 pKa = 3.19VTLIDD300 pKa = 3.82TDD302 pKa = 4.18VISDD306 pKa = 3.63GTYY309 pKa = 8.67GANYY313 pKa = 8.95SAVYY317 pKa = 8.78MWANPDD323 pKa = 3.56PEE325 pKa = 5.03NGVNLVYY332 pKa = 10.88DD333 pKa = 4.14SACTFSNLNSTTNDD347 pKa = 2.79LFFYY351 pKa = 10.44GNNGANITAGLAVAQAGDD369 pKa = 4.06RR370 pKa = 11.84IYY372 pKa = 11.46NSLQAAIDD380 pKa = 4.12DD381 pKa = 4.35DD382 pKa = 4.48NEE384 pKa = 4.33EE385 pKa = 4.01ITLVGDD391 pKa = 3.42TKK393 pKa = 11.34EE394 pKa = 4.07NVVVDD399 pKa = 4.52GQTVTLDD406 pKa = 3.77LNGKK410 pKa = 6.77TVTNVDD416 pKa = 3.23DD417 pKa = 3.5HH418 pKa = 6.77TITVKK423 pKa = 10.67SGEE426 pKa = 4.21LTVKK430 pKa = 10.76GKK432 pKa = 8.76GTVQNLTHH440 pKa = 6.32QKK442 pKa = 10.27AALAAYY448 pKa = 7.92PGTKK452 pKa = 8.63VTLNGGTFDD461 pKa = 3.78RR462 pKa = 11.84TNEE465 pKa = 4.15TGTGPDD471 pKa = 3.12TSGNNSWYY479 pKa = 9.26TIKK482 pKa = 10.72NHH484 pKa = 6.2GDD486 pKa = 3.24MTIDD490 pKa = 3.39GATVRR495 pKa = 11.84NGGGFSSCINNGWYY509 pKa = 10.05SGSAQGTPGNNDD521 pKa = 3.29LAVYY525 pKa = 10.48AGTNAKK531 pKa = 8.59LTIVSGKK538 pKa = 9.79IDD540 pKa = 3.59GGLNAVKK547 pKa = 10.5NDD549 pKa = 3.86DD550 pKa = 3.85YY551 pKa = 12.05GEE553 pKa = 4.42LLVQGGEE560 pKa = 4.33LTNSVQATILNWNIATIEE578 pKa = 4.16KK579 pKa = 9.02GTFSATSDD587 pKa = 3.24AKK589 pKa = 11.01AVILNGKK596 pKa = 9.18IDD598 pKa = 3.43GTMDD602 pKa = 3.28KK603 pKa = 11.03GQLTISDD610 pKa = 4.06GTFSGPVGIQMMSGGVGIGDD630 pKa = 3.48VTITGGTFNISSDD643 pKa = 4.04NIIDD647 pKa = 4.22PLTGTEE653 pKa = 4.17TNVQVSGGSFSDD665 pKa = 4.12DD666 pKa = 2.96SVLAYY671 pKa = 10.43LVEE674 pKa = 4.46GKK676 pKa = 10.84GLVEE680 pKa = 4.5NGTMFDD686 pKa = 3.52VVDD689 pKa = 3.8VHH691 pKa = 5.86TVTYY695 pKa = 9.09TFNDD699 pKa = 3.14TTYY702 pKa = 9.8IEE704 pKa = 4.53KK705 pKa = 10.73VISGQTATQNTIPALPTLPEE725 pKa = 4.01GCEE728 pKa = 3.79YY729 pKa = 11.26DD730 pKa = 3.44FGEE733 pKa = 4.19WDD735 pKa = 3.08KK736 pKa = 10.37TAPVTEE742 pKa = 4.42DD743 pKa = 2.98VSVSVKK749 pKa = 11.1AEE751 pKa = 3.87ITSLTVAIGTSTVDD765 pKa = 3.34GVTTLTATVDD775 pKa = 3.64STASGTPAYY784 pKa = 8.03TWSSGEE790 pKa = 3.95ATEE793 pKa = 5.49SITPSVGGTYY803 pKa = 10.62DD804 pKa = 3.25VTVTLTVTSGDD815 pKa = 3.44VTVTGKK821 pKa = 10.8ANASVTYY828 pKa = 9.02TPSEE832 pKa = 4.32SGSTTTTTTNPDD844 pKa = 2.85GSTTTVEE851 pKa = 4.1TDD853 pKa = 2.56SDD855 pKa = 4.23GSVTEE860 pKa = 4.45TTTKK864 pKa = 10.63TEE866 pKa = 3.93TTEE869 pKa = 3.96TGNEE873 pKa = 3.66ITTVTTTGSDD883 pKa = 3.36PSGSQTTSSTEE894 pKa = 3.81VTIEE898 pKa = 3.99AASGTSGSVSLPEE911 pKa = 3.92EE912 pKa = 4.36DD913 pKa = 3.6VNSVIEE919 pKa = 4.56SIASAGTTTNTISVNIGEE937 pKa = 4.66NDD939 pKa = 3.65TVTMGSAAISAIAEE953 pKa = 4.24SNAVLEE959 pKa = 4.38ISNEE963 pKa = 3.94NATVVADD970 pKa = 3.66GDD972 pKa = 4.19VIEE975 pKa = 4.64TLSTGDD981 pKa = 3.13VSITVGSATHH991 pKa = 7.22DD992 pKa = 3.74NLNEE996 pKa = 4.1SQQQSVPEE1004 pKa = 3.81NSTIVEE1010 pKa = 4.23LSATVGTSPVHH1021 pKa = 6.39DD1022 pKa = 4.39LNGTVEE1028 pKa = 4.09VTMTYY1033 pKa = 9.25TLPSGIDD1040 pKa = 3.35ADD1042 pKa = 4.2DD1043 pKa = 4.62VIVFYY1048 pKa = 11.37VDD1050 pKa = 4.06DD1051 pKa = 5.87DD1052 pKa = 4.5GALHH1056 pKa = 6.49QMRR1059 pKa = 11.84TVYY1062 pKa = 10.91AEE1064 pKa = 4.86GILTFYY1070 pKa = 7.38TTHH1073 pKa = 6.24FSYY1076 pKa = 11.22YY1077 pKa = 9.83FVGDD1081 pKa = 3.17RR1082 pKa = 11.84SMIPSVDD1089 pKa = 3.84PEE1091 pKa = 4.26PSPGEE1096 pKa = 3.9DD1097 pKa = 3.45DD1098 pKa = 3.78QPVVVPPVDD1107 pKa = 5.25DD1108 pKa = 5.45DD1109 pKa = 4.75DD1110 pKa = 7.13DD1111 pKa = 4.39YY1112 pKa = 12.26VPLPPIVDD1120 pKa = 3.66TDD1122 pKa = 3.89GSGSSGGDD1130 pKa = 3.11DD1131 pKa = 3.19TVKK1134 pKa = 10.49IVACAAAAVVAALMAAFLIISRR1156 pKa = 11.84RR1157 pKa = 11.84DD1158 pKa = 3.09
Molecular weight: 119.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B9T8U7|A0A6B9T8U7_9ARCH Abortive phage infection protein OS=Methanomassiliicoccaceae archaeon DOK OX=1535962 GN=JS82_00450 PE=4 SV=1
MM1 pKa = 7.67SSTKK5 pKa = 10.4APAMKK10 pKa = 9.91QRR12 pKa = 11.84LNKK15 pKa = 9.84KK16 pKa = 8.11VNQNRR21 pKa = 11.84RR22 pKa = 11.84VPAWVMLRR30 pKa = 11.84TNRR33 pKa = 11.84QFLRR37 pKa = 11.84HH38 pKa = 5.85PKK40 pKa = 8.59RR41 pKa = 11.84RR42 pKa = 11.84SWRR45 pKa = 11.84MSKK48 pKa = 10.58LKK50 pKa = 10.36EE51 pKa = 3.79
MM1 pKa = 7.67SSTKK5 pKa = 10.4APAMKK10 pKa = 9.91QRR12 pKa = 11.84LNKK15 pKa = 9.84KK16 pKa = 8.11VNQNRR21 pKa = 11.84RR22 pKa = 11.84VPAWVMLRR30 pKa = 11.84TNRR33 pKa = 11.84QFLRR37 pKa = 11.84HH38 pKa = 5.85PKK40 pKa = 8.59RR41 pKa = 11.84RR42 pKa = 11.84SWRR45 pKa = 11.84MSKK48 pKa = 10.58LKK50 pKa = 10.36EE51 pKa = 3.79
Molecular weight: 6.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
554497 |
27 |
3418 |
302.2 |
33.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.112 ± 0.066 | 1.607 ± 0.029 |
7.116 ± 0.052 | 6.834 ± 0.062 |
3.592 ± 0.038 | 8.296 ± 0.065 |
1.636 ± 0.021 | 6.402 ± 0.05 |
4.328 ± 0.06 | 8.031 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.498 ± 0.038 | 3.303 ± 0.055 |
4.029 ± 0.041 | 2.278 ± 0.029 |
5.948 ± 0.091 | 6.531 ± 0.058 |
5.713 ± 0.095 | 8.365 ± 0.059 |
0.964 ± 0.021 | 3.416 ± 0.047 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
