
Beihai mantis shrimp virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
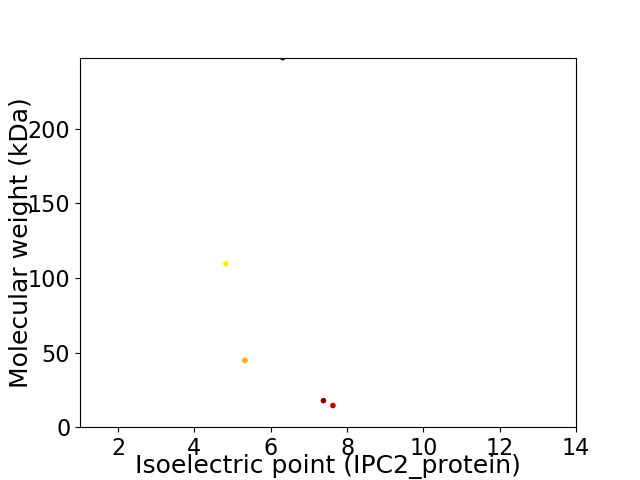
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJI5|A0A1L3KJI5_9VIRU Uncharacterized protein OS=Beihai mantis shrimp virus 2 OX=1922429 PE=4 SV=1
MM1 pKa = 7.72DD2 pKa = 5.36VIHH5 pKa = 6.03THH7 pKa = 4.59SAKK10 pKa = 10.12PVRR13 pKa = 11.84AVTFNNSQATPVRR26 pKa = 11.84VLNMPDD32 pKa = 2.72ITTLQSEE39 pKa = 3.98IDD41 pKa = 3.64YY42 pKa = 10.9LKK44 pKa = 10.75RR45 pKa = 11.84QILALQGDD53 pKa = 4.87FSQISSINEE62 pKa = 3.31ILAQGLATQKK72 pKa = 11.18AFLQRR77 pKa = 11.84LSNLAATARR86 pKa = 11.84NNSFTNTNQINQASAIPPGDD106 pKa = 3.59STLGKK111 pKa = 9.35PSAISLVYY119 pKa = 10.02FLLHH123 pKa = 6.7SMGSSDD129 pKa = 3.78IKK131 pKa = 10.96LSSTEE136 pKa = 3.59IKK138 pKa = 10.91NFLSQDD144 pKa = 3.28LNLCKK149 pKa = 9.64FTNEE153 pKa = 3.81IFSFGPEE160 pKa = 3.8ITPKK164 pKa = 10.45AIEE167 pKa = 4.21FTVPEE172 pKa = 4.12PTLEE176 pKa = 4.18EE177 pKa = 4.21IYY179 pKa = 10.64LVSIEE184 pKa = 4.62PGKK187 pKa = 10.47DD188 pKa = 3.23DD189 pKa = 4.42QSVVLYY195 pKa = 9.98KK196 pKa = 10.21FNIISNTWEE205 pKa = 4.11TEE207 pKa = 4.05VLGTCAIDD215 pKa = 3.62FSNFSVTNTQGILQCAYY232 pKa = 9.11GQFVATSTSNVDD244 pKa = 4.06LYY246 pKa = 11.43SQNALLNYY254 pKa = 8.11TFVRR258 pKa = 11.84ALEE261 pKa = 4.42SYY263 pKa = 9.58TLLLEE268 pKa = 4.14EE269 pKa = 5.56LIEE272 pKa = 4.42SALSSVSQTKK282 pKa = 10.34LNPIVEE288 pKa = 4.17EE289 pKa = 3.85ALRR292 pKa = 11.84IEE294 pKa = 4.0WLLLQFQEE302 pKa = 5.17SLLSMQANGIFYY314 pKa = 10.78VEE316 pKa = 4.27STSDD320 pKa = 3.61YY321 pKa = 10.02VSEE324 pKa = 4.51INFSDD329 pKa = 3.54VTKK332 pKa = 10.23PLSTVSCQSSINYY345 pKa = 9.61SVTPEE350 pKa = 4.41GYY352 pKa = 9.3VGSVSAEE359 pKa = 3.7PAFEE363 pKa = 4.52PEE365 pKa = 4.03QQEE368 pKa = 4.94GYY370 pKa = 10.92SDD372 pKa = 3.66IKK374 pKa = 11.33NLDD377 pKa = 3.74DD378 pKa = 4.72FLLTVTTHH386 pKa = 6.52SEE388 pKa = 4.44LEE390 pKa = 3.75FHH392 pKa = 7.01EE393 pKa = 4.99YY394 pKa = 9.44IVHH397 pKa = 6.79EE398 pKa = 4.29DD399 pKa = 3.3HH400 pKa = 7.87DD401 pKa = 4.08VLEE404 pKa = 5.6DD405 pKa = 3.64YY406 pKa = 11.3RR407 pKa = 11.84KK408 pKa = 8.35MTVSQEE414 pKa = 3.92KK415 pKa = 10.13PNLHH419 pKa = 6.88HH420 pKa = 6.74YY421 pKa = 10.84VDD423 pKa = 3.99IKK425 pKa = 10.88ISGSSFPGMEE435 pKa = 4.36VDD437 pKa = 4.06VQPCTVDD444 pKa = 3.14RR445 pKa = 11.84DD446 pKa = 3.68GDD448 pKa = 3.57ISQVSFDD455 pKa = 3.49TTYY458 pKa = 11.33NADD461 pKa = 2.99GSYY464 pKa = 10.36QAVASVWVGARR475 pKa = 11.84GLFYY479 pKa = 10.47KK480 pKa = 10.86YY481 pKa = 10.25EE482 pKa = 4.17VDD484 pKa = 3.97GKK486 pKa = 8.36TSQSGRR492 pKa = 11.84YY493 pKa = 8.79KK494 pKa = 9.87EE495 pKa = 4.23NKK497 pKa = 8.71VLSEE501 pKa = 4.09NEE503 pKa = 3.63QVEE506 pKa = 4.59IVTTASLEE514 pKa = 4.0GDD516 pKa = 4.78KK517 pKa = 10.4IDD519 pKa = 4.49CAQKK523 pKa = 8.59IHH525 pKa = 6.91GYY527 pKa = 10.07ALEE530 pKa = 4.34FNSFNSVVKK539 pKa = 10.54ISGFKK544 pKa = 10.01VRR546 pKa = 11.84YY547 pKa = 6.5PKK549 pKa = 10.54PPVFSFVPSFDD560 pKa = 3.59KK561 pKa = 11.19QQTEE565 pKa = 3.92LMAITFKK572 pKa = 10.13NTYY575 pKa = 9.6EE576 pKa = 4.35SIKK579 pKa = 10.05LTAEE583 pKa = 4.08YY584 pKa = 9.52EE585 pKa = 4.19QLSARR590 pKa = 11.84LEE592 pKa = 4.13ALGKK596 pKa = 9.83ICSPTLLGAFGGLFGTASNFAKK618 pKa = 10.25SVKK621 pKa = 9.76AACTLQKK628 pKa = 10.34LASGFYY634 pKa = 10.43GLEE637 pKa = 3.69EE638 pKa = 4.24LVQGRR643 pKa = 11.84YY644 pKa = 9.12IGAILGFAGASTASLTNKK662 pKa = 9.93LVRR665 pKa = 11.84KK666 pKa = 10.12GSFEE670 pKa = 4.31NKK672 pKa = 9.63DD673 pKa = 3.48DD674 pKa = 5.65LIGHH678 pKa = 5.33VAKK681 pKa = 10.04TIRR684 pKa = 11.84FSKK687 pKa = 10.39YY688 pKa = 9.89LPEE691 pKa = 4.27LNKK694 pKa = 10.4RR695 pKa = 11.84KK696 pKa = 9.97KK697 pKa = 10.28NYY699 pKa = 9.84SNSPFEE705 pKa = 4.3TMSTSDD711 pKa = 3.29NFIAKK716 pKa = 8.9MGAPINQLPGSEE728 pKa = 4.86FITNAFLNVTPEE740 pKa = 3.99DD741 pKa = 3.59KK742 pKa = 10.55SYY744 pKa = 11.09KK745 pKa = 9.9RR746 pKa = 11.84IMKK749 pKa = 9.88IKK751 pKa = 9.09EE752 pKa = 3.64DD753 pKa = 4.27GYY755 pKa = 10.9FPEE758 pKa = 4.6HH759 pKa = 6.53QSIFTNTMFTVGPDD773 pKa = 2.91VDD775 pKa = 4.0TEE777 pKa = 4.28EE778 pKa = 4.9LFVMQCRR785 pKa = 11.84YY786 pKa = 10.11GISDD790 pKa = 3.53GAVNRR795 pKa = 11.84IHH797 pKa = 6.93SFNTGGSSAKK807 pKa = 9.15TVAGPGSSFLIKK819 pKa = 10.06KK820 pKa = 10.22DD821 pKa = 3.37GEE823 pKa = 4.33TSVPYY828 pKa = 10.6LINGKK833 pKa = 8.39IEE835 pKa = 4.13KK836 pKa = 9.12MALLAMDD843 pKa = 5.76GYY845 pKa = 11.21DD846 pKa = 3.03PSLFTEE852 pKa = 4.53EE853 pKa = 4.61MSDD856 pKa = 3.41TDD858 pKa = 5.84LEE860 pKa = 4.83SIDD863 pKa = 4.11NRR865 pKa = 11.84TNMEE869 pKa = 3.76IALSIDD875 pKa = 3.07KK876 pKa = 10.74KK877 pKa = 9.89MKK879 pKa = 9.83FYY881 pKa = 10.53NSRR884 pKa = 11.84VIEE887 pKa = 4.13SSPFQSTPEE896 pKa = 3.91ANRR899 pKa = 11.84DD900 pKa = 3.41ALASLMSGSLFKK912 pKa = 10.93DD913 pKa = 3.94FNYY916 pKa = 10.49SIPNRR921 pKa = 11.84TCQPFADD928 pKa = 3.32ALYY931 pKa = 10.65NNMLKK936 pKa = 10.13SEE938 pKa = 4.08QAPILGGFDD947 pKa = 3.3FNSRR951 pKa = 11.84HH952 pKa = 6.24RR953 pKa = 11.84LTLEE957 pKa = 3.71PMSYY961 pKa = 11.5ADD963 pKa = 6.06LDD965 pKa = 4.1DD966 pKa = 6.55LMLQTYY972 pKa = 10.21RR973 pKa = 11.84GILSFDD979 pKa = 2.88IFVINNII986 pKa = 3.91
MM1 pKa = 7.72DD2 pKa = 5.36VIHH5 pKa = 6.03THH7 pKa = 4.59SAKK10 pKa = 10.12PVRR13 pKa = 11.84AVTFNNSQATPVRR26 pKa = 11.84VLNMPDD32 pKa = 2.72ITTLQSEE39 pKa = 3.98IDD41 pKa = 3.64YY42 pKa = 10.9LKK44 pKa = 10.75RR45 pKa = 11.84QILALQGDD53 pKa = 4.87FSQISSINEE62 pKa = 3.31ILAQGLATQKK72 pKa = 11.18AFLQRR77 pKa = 11.84LSNLAATARR86 pKa = 11.84NNSFTNTNQINQASAIPPGDD106 pKa = 3.59STLGKK111 pKa = 9.35PSAISLVYY119 pKa = 10.02FLLHH123 pKa = 6.7SMGSSDD129 pKa = 3.78IKK131 pKa = 10.96LSSTEE136 pKa = 3.59IKK138 pKa = 10.91NFLSQDD144 pKa = 3.28LNLCKK149 pKa = 9.64FTNEE153 pKa = 3.81IFSFGPEE160 pKa = 3.8ITPKK164 pKa = 10.45AIEE167 pKa = 4.21FTVPEE172 pKa = 4.12PTLEE176 pKa = 4.18EE177 pKa = 4.21IYY179 pKa = 10.64LVSIEE184 pKa = 4.62PGKK187 pKa = 10.47DD188 pKa = 3.23DD189 pKa = 4.42QSVVLYY195 pKa = 9.98KK196 pKa = 10.21FNIISNTWEE205 pKa = 4.11TEE207 pKa = 4.05VLGTCAIDD215 pKa = 3.62FSNFSVTNTQGILQCAYY232 pKa = 9.11GQFVATSTSNVDD244 pKa = 4.06LYY246 pKa = 11.43SQNALLNYY254 pKa = 8.11TFVRR258 pKa = 11.84ALEE261 pKa = 4.42SYY263 pKa = 9.58TLLLEE268 pKa = 4.14EE269 pKa = 5.56LIEE272 pKa = 4.42SALSSVSQTKK282 pKa = 10.34LNPIVEE288 pKa = 4.17EE289 pKa = 3.85ALRR292 pKa = 11.84IEE294 pKa = 4.0WLLLQFQEE302 pKa = 5.17SLLSMQANGIFYY314 pKa = 10.78VEE316 pKa = 4.27STSDD320 pKa = 3.61YY321 pKa = 10.02VSEE324 pKa = 4.51INFSDD329 pKa = 3.54VTKK332 pKa = 10.23PLSTVSCQSSINYY345 pKa = 9.61SVTPEE350 pKa = 4.41GYY352 pKa = 9.3VGSVSAEE359 pKa = 3.7PAFEE363 pKa = 4.52PEE365 pKa = 4.03QQEE368 pKa = 4.94GYY370 pKa = 10.92SDD372 pKa = 3.66IKK374 pKa = 11.33NLDD377 pKa = 3.74DD378 pKa = 4.72FLLTVTTHH386 pKa = 6.52SEE388 pKa = 4.44LEE390 pKa = 3.75FHH392 pKa = 7.01EE393 pKa = 4.99YY394 pKa = 9.44IVHH397 pKa = 6.79EE398 pKa = 4.29DD399 pKa = 3.3HH400 pKa = 7.87DD401 pKa = 4.08VLEE404 pKa = 5.6DD405 pKa = 3.64YY406 pKa = 11.3RR407 pKa = 11.84KK408 pKa = 8.35MTVSQEE414 pKa = 3.92KK415 pKa = 10.13PNLHH419 pKa = 6.88HH420 pKa = 6.74YY421 pKa = 10.84VDD423 pKa = 3.99IKK425 pKa = 10.88ISGSSFPGMEE435 pKa = 4.36VDD437 pKa = 4.06VQPCTVDD444 pKa = 3.14RR445 pKa = 11.84DD446 pKa = 3.68GDD448 pKa = 3.57ISQVSFDD455 pKa = 3.49TTYY458 pKa = 11.33NADD461 pKa = 2.99GSYY464 pKa = 10.36QAVASVWVGARR475 pKa = 11.84GLFYY479 pKa = 10.47KK480 pKa = 10.86YY481 pKa = 10.25EE482 pKa = 4.17VDD484 pKa = 3.97GKK486 pKa = 8.36TSQSGRR492 pKa = 11.84YY493 pKa = 8.79KK494 pKa = 9.87EE495 pKa = 4.23NKK497 pKa = 8.71VLSEE501 pKa = 4.09NEE503 pKa = 3.63QVEE506 pKa = 4.59IVTTASLEE514 pKa = 4.0GDD516 pKa = 4.78KK517 pKa = 10.4IDD519 pKa = 4.49CAQKK523 pKa = 8.59IHH525 pKa = 6.91GYY527 pKa = 10.07ALEE530 pKa = 4.34FNSFNSVVKK539 pKa = 10.54ISGFKK544 pKa = 10.01VRR546 pKa = 11.84YY547 pKa = 6.5PKK549 pKa = 10.54PPVFSFVPSFDD560 pKa = 3.59KK561 pKa = 11.19QQTEE565 pKa = 3.92LMAITFKK572 pKa = 10.13NTYY575 pKa = 9.6EE576 pKa = 4.35SIKK579 pKa = 10.05LTAEE583 pKa = 4.08YY584 pKa = 9.52EE585 pKa = 4.19QLSARR590 pKa = 11.84LEE592 pKa = 4.13ALGKK596 pKa = 9.83ICSPTLLGAFGGLFGTASNFAKK618 pKa = 10.25SVKK621 pKa = 9.76AACTLQKK628 pKa = 10.34LASGFYY634 pKa = 10.43GLEE637 pKa = 3.69EE638 pKa = 4.24LVQGRR643 pKa = 11.84YY644 pKa = 9.12IGAILGFAGASTASLTNKK662 pKa = 9.93LVRR665 pKa = 11.84KK666 pKa = 10.12GSFEE670 pKa = 4.31NKK672 pKa = 9.63DD673 pKa = 3.48DD674 pKa = 5.65LIGHH678 pKa = 5.33VAKK681 pKa = 10.04TIRR684 pKa = 11.84FSKK687 pKa = 10.39YY688 pKa = 9.89LPEE691 pKa = 4.27LNKK694 pKa = 10.4RR695 pKa = 11.84KK696 pKa = 9.97KK697 pKa = 10.28NYY699 pKa = 9.84SNSPFEE705 pKa = 4.3TMSTSDD711 pKa = 3.29NFIAKK716 pKa = 8.9MGAPINQLPGSEE728 pKa = 4.86FITNAFLNVTPEE740 pKa = 3.99DD741 pKa = 3.59KK742 pKa = 10.55SYY744 pKa = 11.09KK745 pKa = 9.9RR746 pKa = 11.84IMKK749 pKa = 9.88IKK751 pKa = 9.09EE752 pKa = 3.64DD753 pKa = 4.27GYY755 pKa = 10.9FPEE758 pKa = 4.6HH759 pKa = 6.53QSIFTNTMFTVGPDD773 pKa = 2.91VDD775 pKa = 4.0TEE777 pKa = 4.28EE778 pKa = 4.9LFVMQCRR785 pKa = 11.84YY786 pKa = 10.11GISDD790 pKa = 3.53GAVNRR795 pKa = 11.84IHH797 pKa = 6.93SFNTGGSSAKK807 pKa = 9.15TVAGPGSSFLIKK819 pKa = 10.06KK820 pKa = 10.22DD821 pKa = 3.37GEE823 pKa = 4.33TSVPYY828 pKa = 10.6LINGKK833 pKa = 8.39IEE835 pKa = 4.13KK836 pKa = 9.12MALLAMDD843 pKa = 5.76GYY845 pKa = 11.21DD846 pKa = 3.03PSLFTEE852 pKa = 4.53EE853 pKa = 4.61MSDD856 pKa = 3.41TDD858 pKa = 5.84LEE860 pKa = 4.83SIDD863 pKa = 4.11NRR865 pKa = 11.84TNMEE869 pKa = 3.76IALSIDD875 pKa = 3.07KK876 pKa = 10.74KK877 pKa = 9.89MKK879 pKa = 9.83FYY881 pKa = 10.53NSRR884 pKa = 11.84VIEE887 pKa = 4.13SSPFQSTPEE896 pKa = 3.91ANRR899 pKa = 11.84DD900 pKa = 3.41ALASLMSGSLFKK912 pKa = 10.93DD913 pKa = 3.94FNYY916 pKa = 10.49SIPNRR921 pKa = 11.84TCQPFADD928 pKa = 3.32ALYY931 pKa = 10.65NNMLKK936 pKa = 10.13SEE938 pKa = 4.08QAPILGGFDD947 pKa = 3.3FNSRR951 pKa = 11.84HH952 pKa = 6.24RR953 pKa = 11.84LTLEE957 pKa = 3.71PMSYY961 pKa = 11.5ADD963 pKa = 6.06LDD965 pKa = 4.1DD966 pKa = 6.55LMLQTYY972 pKa = 10.21RR973 pKa = 11.84GILSFDD979 pKa = 2.88IFVINNII986 pKa = 3.91
Molecular weight: 109.52 kDa
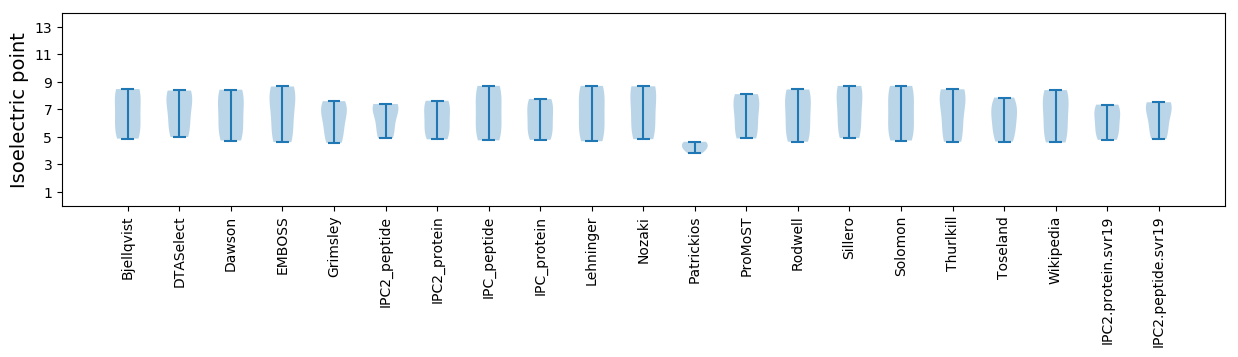
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJJ1|A0A1L3KJJ1_9VIRU Uncharacterized protein OS=Beihai mantis shrimp virus 2 OX=1922429 PE=4 SV=1
MM1 pKa = 7.67SSTNAYY7 pKa = 9.64VLNLRR12 pKa = 11.84AKK14 pKa = 9.51DD15 pKa = 3.48AKK17 pKa = 11.04LEE19 pKa = 4.06IVKK22 pKa = 10.3DD23 pKa = 3.65FTMVPGRR30 pKa = 11.84DD31 pKa = 3.78PLEE34 pKa = 4.46HH35 pKa = 6.57RR36 pKa = 11.84CKK38 pKa = 10.4ATICVKK44 pKa = 10.3GKK46 pKa = 9.81HH47 pKa = 5.33FEE49 pKa = 4.38VVQSAQSKK57 pKa = 8.31KK58 pKa = 10.36AAYY61 pKa = 8.18EE62 pKa = 3.93KK63 pKa = 10.91AAGEE67 pKa = 4.4LLQLWPYY74 pKa = 10.11VEE76 pKa = 4.69EE77 pKa = 4.24VLNEE81 pKa = 3.87QLNEE85 pKa = 4.09EE86 pKa = 4.65EE87 pKa = 4.76LTVLDD92 pKa = 3.76INVVKK97 pKa = 9.04TNKK100 pKa = 8.99MFSVYY105 pKa = 10.16RR106 pKa = 11.84GSTQKK111 pKa = 10.55NFKK114 pKa = 10.07KK115 pKa = 10.63KK116 pKa = 10.36SDD118 pKa = 3.48LLTYY122 pKa = 10.59LEE124 pKa = 5.08NILL127 pKa = 4.55
MM1 pKa = 7.67SSTNAYY7 pKa = 9.64VLNLRR12 pKa = 11.84AKK14 pKa = 9.51DD15 pKa = 3.48AKK17 pKa = 11.04LEE19 pKa = 4.06IVKK22 pKa = 10.3DD23 pKa = 3.65FTMVPGRR30 pKa = 11.84DD31 pKa = 3.78PLEE34 pKa = 4.46HH35 pKa = 6.57RR36 pKa = 11.84CKK38 pKa = 10.4ATICVKK44 pKa = 10.3GKK46 pKa = 9.81HH47 pKa = 5.33FEE49 pKa = 4.38VVQSAQSKK57 pKa = 8.31KK58 pKa = 10.36AAYY61 pKa = 8.18EE62 pKa = 3.93KK63 pKa = 10.91AAGEE67 pKa = 4.4LLQLWPYY74 pKa = 10.11VEE76 pKa = 4.69EE77 pKa = 4.24VLNEE81 pKa = 3.87QLNEE85 pKa = 4.09EE86 pKa = 4.65EE87 pKa = 4.76LTVLDD92 pKa = 3.76INVVKK97 pKa = 9.04TNKK100 pKa = 8.99MFSVYY105 pKa = 10.16RR106 pKa = 11.84GSTQKK111 pKa = 10.55NFKK114 pKa = 10.07KK115 pKa = 10.63KK116 pKa = 10.36SDD118 pKa = 3.48LLTYY122 pKa = 10.59LEE124 pKa = 5.08NILL127 pKa = 4.55
Molecular weight: 14.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3860 |
127 |
2185 |
772.0 |
86.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.658 ± 0.248 | 1.58 ± 0.374 |
5.984 ± 0.287 | 6.088 ± 0.375 |
5.855 ± 0.331 | 4.948 ± 0.451 |
2.021 ± 0.251 | 5.881 ± 0.329 |
6.917 ± 0.59 | 9.145 ± 0.36 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.684 ± 0.24 | 5.648 ± 0.511 |
5.181 ± 0.49 | 3.497 ± 0.282 |
3.886 ± 0.586 | 7.124 ± 1.259 |
6.969 ± 1.001 | 6.477 ± 0.465 |
0.622 ± 0.13 | 3.834 ± 0.255 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
