
Chamois faeces associated circular DNA virus 1
Taxonomy: Viruses; unclassified viruses
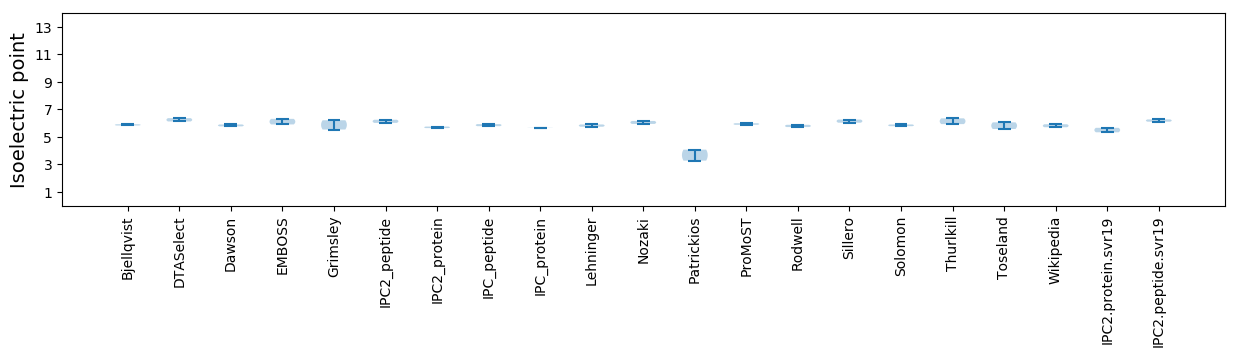
Average proteome isoelectric point is 5.5
Get precalculated fractions of proteins
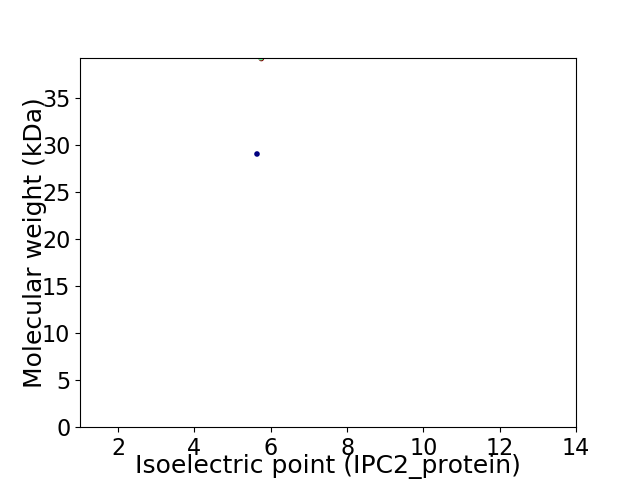
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A168MFW5|A0A168MFW5_9VIRU Replication associated protein OS=Chamois faeces associated circular DNA virus 1 OX=1843766 PE=4 SV=1
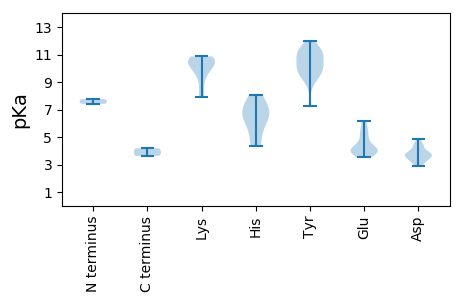
MM1 pKa = 7.41LTQDD5 pKa = 3.76VPIISNPAGDD15 pKa = 4.42DD16 pKa = 3.61GGVLIQHH23 pKa = 6.75RR24 pKa = 11.84EE25 pKa = 3.96YY26 pKa = 11.01LCDD29 pKa = 3.85VISTGSAFAIQNQITINPGNPACFPWLSTIAQNFTQYY66 pKa = 11.03KK67 pKa = 9.15LEE69 pKa = 3.82GMMFNYY75 pKa = 10.03VSTSGALSTTQALGEE90 pKa = 4.3IIMAVDD96 pKa = 3.68YY97 pKa = 11.38NPAGPNFSSKK107 pKa = 8.37QQMLNQVFAVSKK119 pKa = 10.69VPSEE123 pKa = 4.42DD124 pKa = 3.08AVCPIEE130 pKa = 5.52CDD132 pKa = 3.39PKK134 pKa = 10.08QTGTGDD140 pKa = 3.56LLYY143 pKa = 10.5TRR145 pKa = 11.84GATIPIGQDD154 pKa = 2.86PRR156 pKa = 11.84FYY158 pKa = 11.16DD159 pKa = 3.66CGTFSLATQGQTAGVTLGEE178 pKa = 3.95LWITYY183 pKa = 7.22QVQLYY188 pKa = 9.34KK189 pKa = 10.42PQSLAQLPSLAAPNIFNVRR208 pKa = 11.84SSPANTALVSLMGGSTGFVSTGEE231 pKa = 4.08MVPTIPASSSSMTFTTFYY249 pKa = 10.51PAGTSFGLFCTVTGAGMTAPPNITPGTGTILAGGFPNAASAVRR292 pKa = 11.84VGAVTEE298 pKa = 4.04ASFAGRR304 pKa = 11.84FQCTNSVQQWTLVGAFGLTGGNVQIVGWIRR334 pKa = 11.84RR335 pKa = 11.84TSNTSSTWSFLTILRR350 pKa = 11.84VILSLLSTIRR360 pKa = 11.84TSRR363 pKa = 11.84SAMMFRR369 pKa = 11.84VGG371 pKa = 4.18
MM1 pKa = 7.41LTQDD5 pKa = 3.76VPIISNPAGDD15 pKa = 4.42DD16 pKa = 3.61GGVLIQHH23 pKa = 6.75RR24 pKa = 11.84EE25 pKa = 3.96YY26 pKa = 11.01LCDD29 pKa = 3.85VISTGSAFAIQNQITINPGNPACFPWLSTIAQNFTQYY66 pKa = 11.03KK67 pKa = 9.15LEE69 pKa = 3.82GMMFNYY75 pKa = 10.03VSTSGALSTTQALGEE90 pKa = 4.3IIMAVDD96 pKa = 3.68YY97 pKa = 11.38NPAGPNFSSKK107 pKa = 8.37QQMLNQVFAVSKK119 pKa = 10.69VPSEE123 pKa = 4.42DD124 pKa = 3.08AVCPIEE130 pKa = 5.52CDD132 pKa = 3.39PKK134 pKa = 10.08QTGTGDD140 pKa = 3.56LLYY143 pKa = 10.5TRR145 pKa = 11.84GATIPIGQDD154 pKa = 2.86PRR156 pKa = 11.84FYY158 pKa = 11.16DD159 pKa = 3.66CGTFSLATQGQTAGVTLGEE178 pKa = 3.95LWITYY183 pKa = 7.22QVQLYY188 pKa = 9.34KK189 pKa = 10.42PQSLAQLPSLAAPNIFNVRR208 pKa = 11.84SSPANTALVSLMGGSTGFVSTGEE231 pKa = 4.08MVPTIPASSSSMTFTTFYY249 pKa = 10.51PAGTSFGLFCTVTGAGMTAPPNITPGTGTILAGGFPNAASAVRR292 pKa = 11.84VGAVTEE298 pKa = 4.04ASFAGRR304 pKa = 11.84FQCTNSVQQWTLVGAFGLTGGNVQIVGWIRR334 pKa = 11.84RR335 pKa = 11.84TSNTSSTWSFLTILRR350 pKa = 11.84VILSLLSTIRR360 pKa = 11.84TSRR363 pKa = 11.84SAMMFRR369 pKa = 11.84VGG371 pKa = 4.18
Molecular weight: 39.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168MFW5|A0A168MFW5_9VIRU Replication associated protein OS=Chamois faeces associated circular DNA virus 1 OX=1843766 PE=4 SV=1
MM1 pKa = 7.76EE2 pKa = 5.08FPDD5 pKa = 4.05YY6 pKa = 11.32LKK8 pKa = 10.86GDD10 pKa = 4.04PLSPVYY16 pKa = 10.45HH17 pKa = 6.53PNIKK21 pKa = 9.7KK22 pKa = 10.17CHH24 pKa = 6.33DD25 pKa = 3.2VSGWLKK31 pKa = 10.74YY32 pKa = 10.03ISKK35 pKa = 10.64DD36 pKa = 3.36GEE38 pKa = 4.19FVDD41 pKa = 3.93MTGKK45 pKa = 10.49FNILDD50 pKa = 3.76YY51 pKa = 11.2EE52 pKa = 4.18VGKK55 pKa = 10.01RR56 pKa = 11.84QKK58 pKa = 10.7AYY60 pKa = 9.86QDD62 pKa = 3.36HH63 pKa = 7.22KK64 pKa = 10.75FTEE67 pKa = 3.93DD68 pKa = 2.88WIRR71 pKa = 11.84RR72 pKa = 11.84KK73 pKa = 9.98NYY75 pKa = 9.7KK76 pKa = 10.01EE77 pKa = 3.85VVFPVLLKK85 pKa = 8.09TTTGRR90 pKa = 11.84EE91 pKa = 3.94YY92 pKa = 11.34RR93 pKa = 11.84MNAPDD98 pKa = 3.76ASVKK102 pKa = 9.16QRR104 pKa = 11.84HH105 pKa = 4.9WWIVAPPNSGKK116 pKa = 7.93TRR118 pKa = 11.84WLQDD122 pKa = 3.0TFDD125 pKa = 3.75GQRR128 pKa = 11.84VYY130 pKa = 11.11VPSKK134 pKa = 10.6GEE136 pKa = 3.98YY137 pKa = 9.42PFEE140 pKa = 6.05RR141 pKa = 11.84YY142 pKa = 9.52DD143 pKa = 3.39GQSIVVYY150 pKa = 10.01DD151 pKa = 4.47DD152 pKa = 3.78RR153 pKa = 11.84DD154 pKa = 3.9SITFEE159 pKa = 3.85EE160 pKa = 4.93FSNVTGVYY168 pKa = 10.14KK169 pKa = 10.46IEE171 pKa = 3.79THH173 pKa = 6.1VYY175 pKa = 8.97GKK177 pKa = 10.73ARR179 pKa = 11.84FVNVFWPEE187 pKa = 3.54HH188 pKa = 4.33QARR191 pKa = 11.84NVIVLSNKK199 pKa = 9.58TIEE202 pKa = 4.2QQCGDD207 pKa = 3.49DD208 pKa = 3.72SYY210 pKa = 11.98RR211 pKa = 11.84MKK213 pKa = 10.76KK214 pKa = 10.18RR215 pKa = 11.84FIQISGAALCPPHH228 pKa = 7.39DD229 pKa = 4.49SSDD232 pKa = 4.35DD233 pKa = 3.64EE234 pKa = 6.14DD235 pKa = 4.42EE236 pKa = 5.14VADD239 pKa = 4.83CPPDD243 pKa = 3.36EE244 pKa = 5.48HH245 pKa = 8.05GFASS249 pKa = 3.63
MM1 pKa = 7.76EE2 pKa = 5.08FPDD5 pKa = 4.05YY6 pKa = 11.32LKK8 pKa = 10.86GDD10 pKa = 4.04PLSPVYY16 pKa = 10.45HH17 pKa = 6.53PNIKK21 pKa = 9.7KK22 pKa = 10.17CHH24 pKa = 6.33DD25 pKa = 3.2VSGWLKK31 pKa = 10.74YY32 pKa = 10.03ISKK35 pKa = 10.64DD36 pKa = 3.36GEE38 pKa = 4.19FVDD41 pKa = 3.93MTGKK45 pKa = 10.49FNILDD50 pKa = 3.76YY51 pKa = 11.2EE52 pKa = 4.18VGKK55 pKa = 10.01RR56 pKa = 11.84QKK58 pKa = 10.7AYY60 pKa = 9.86QDD62 pKa = 3.36HH63 pKa = 7.22KK64 pKa = 10.75FTEE67 pKa = 3.93DD68 pKa = 2.88WIRR71 pKa = 11.84RR72 pKa = 11.84KK73 pKa = 9.98NYY75 pKa = 9.7KK76 pKa = 10.01EE77 pKa = 3.85VVFPVLLKK85 pKa = 8.09TTTGRR90 pKa = 11.84EE91 pKa = 3.94YY92 pKa = 11.34RR93 pKa = 11.84MNAPDD98 pKa = 3.76ASVKK102 pKa = 9.16QRR104 pKa = 11.84HH105 pKa = 4.9WWIVAPPNSGKK116 pKa = 7.93TRR118 pKa = 11.84WLQDD122 pKa = 3.0TFDD125 pKa = 3.75GQRR128 pKa = 11.84VYY130 pKa = 11.11VPSKK134 pKa = 10.6GEE136 pKa = 3.98YY137 pKa = 9.42PFEE140 pKa = 6.05RR141 pKa = 11.84YY142 pKa = 9.52DD143 pKa = 3.39GQSIVVYY150 pKa = 10.01DD151 pKa = 4.47DD152 pKa = 3.78RR153 pKa = 11.84DD154 pKa = 3.9SITFEE159 pKa = 3.85EE160 pKa = 4.93FSNVTGVYY168 pKa = 10.14KK169 pKa = 10.46IEE171 pKa = 3.79THH173 pKa = 6.1VYY175 pKa = 8.97GKK177 pKa = 10.73ARR179 pKa = 11.84FVNVFWPEE187 pKa = 3.54HH188 pKa = 4.33QARR191 pKa = 11.84NVIVLSNKK199 pKa = 9.58TIEE202 pKa = 4.2QQCGDD207 pKa = 3.49DD208 pKa = 3.72SYY210 pKa = 11.98RR211 pKa = 11.84MKK213 pKa = 10.76KK214 pKa = 10.18RR215 pKa = 11.84FIQISGAALCPPHH228 pKa = 7.39DD229 pKa = 4.49SSDD232 pKa = 4.35DD233 pKa = 3.64EE234 pKa = 6.14DD235 pKa = 4.42EE236 pKa = 5.14VADD239 pKa = 4.83CPPDD243 pKa = 3.36EE244 pKa = 5.48HH245 pKa = 8.05GFASS249 pKa = 3.63
Molecular weight: 29.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
620 |
249 |
371 |
310.0 |
34.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.774 ± 1.669 | 1.774 ± 0.102 |
5.323 ± 2.369 | 3.871 ± 1.546 |
5.323 ± 0.062 | 8.387 ± 1.43 |
1.452 ± 1.066 | 5.645 ± 0.5 |
4.032 ± 2.421 | 5.968 ± 1.424 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.419 ± 0.492 | 4.194 ± 0.35 |
6.129 ± 0.064 | 5.161 ± 0.693 |
4.194 ± 0.865 | 8.065 ± 1.235 |
8.387 ± 2.402 | 7.419 ± 0.614 |
1.774 ± 0.385 | 3.71 ± 1.158 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
