
Sphingomonas sp. Leaf357
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas; unclassified Sphingomonas
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
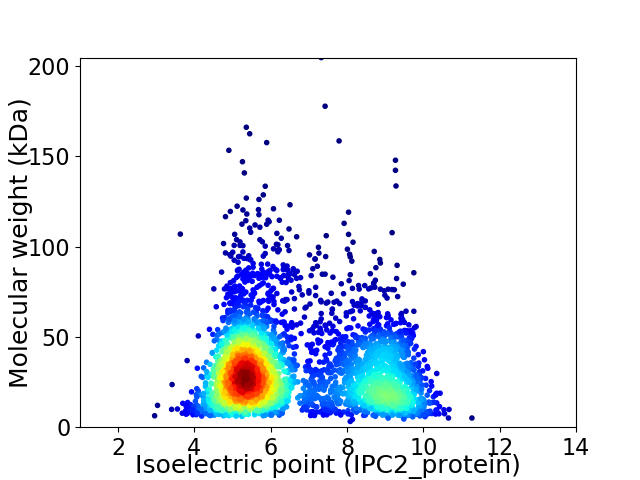
Virtual 2D-PAGE plot for 3524 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q5QXU7|A0A0Q5QXU7_9SPHN PEP-CTERM domain-containing protein OS=Sphingomonas sp. Leaf357 OX=1736350 GN=ASG11_07965 PE=4 SV=1
MM1 pKa = 7.2NLAGVSYY8 pKa = 11.02YY9 pKa = 9.06GTQYY13 pKa = 11.15PFLDD17 pKa = 3.38RR18 pKa = 11.84FKK20 pKa = 10.64TSSEE24 pKa = 3.56WMVQGVNGQPIGALSLDD41 pKa = 3.48ANGYY45 pKa = 5.06PTAMPTGATNIFTMVGLDD63 pKa = 3.6PVALEE68 pKa = 4.02TSDD71 pKa = 4.1IYY73 pKa = 11.53VITYY77 pKa = 9.47KK78 pKa = 10.99GNASINILGGTILSSEE94 pKa = 4.15PGKK97 pKa = 9.24ITFRR101 pKa = 11.84FTGEE105 pKa = 4.03GSSTAINVSAIDD117 pKa = 3.73PANPLSDD124 pKa = 3.36MHH126 pKa = 6.97VVRR129 pKa = 11.84QDD131 pKa = 3.17QVALYY136 pKa = 10.03EE137 pKa = 4.33SGEE140 pKa = 4.07IFNPAFLDD148 pKa = 4.24KK149 pKa = 10.82ASQWSEE155 pKa = 3.69LRR157 pKa = 11.84YY158 pKa = 9.41MDD160 pKa = 4.85WGNTNNNTLTSWDD173 pKa = 3.42QRR175 pKa = 11.84TTLTSASWSNISSTHH190 pKa = 5.03SVPIEE195 pKa = 3.92IMVALANKK203 pKa = 8.67TNTDD207 pKa = 2.64MWINIPAAADD217 pKa = 3.23DD218 pKa = 4.23DD219 pKa = 4.98YY220 pKa = 11.7IRR222 pKa = 11.84KK223 pKa = 6.93TLSYY227 pKa = 11.14VRR229 pKa = 11.84DD230 pKa = 3.56NLAAGLSVKK239 pKa = 10.15VEE241 pKa = 4.02YY242 pKa = 11.14SNEE245 pKa = 3.31VWNWGFQQASYY256 pKa = 9.37AQTMGAQMFGTDD268 pKa = 4.09LNGNGQIDD276 pKa = 3.74RR277 pKa = 11.84GTSEE281 pKa = 4.43EE282 pKa = 4.57PGTAYY287 pKa = 10.33LQYY290 pKa = 9.98YY291 pKa = 8.72GYY293 pKa = 10.67RR294 pKa = 11.84AAQVAAVANSVFASSDD310 pKa = 3.29PARR313 pKa = 11.84LQNVLSTQTGYY324 pKa = 11.29LGLEE328 pKa = 4.02TYY330 pKa = 10.45IFDD333 pKa = 4.43GVQSAALGSVASLFDD348 pKa = 3.41NYY350 pKa = 11.28AITTYY355 pKa = 10.67FGLDD359 pKa = 3.23SVDD362 pKa = 3.39QATLVSWAHH371 pKa = 6.55AGQAGIDD378 pKa = 3.45AAFAGLNASLNDD390 pKa = 3.5SLRR393 pKa = 11.84VYY395 pKa = 10.5VYY397 pKa = 9.57QQAVAARR404 pKa = 11.84NGLSLVAYY412 pKa = 9.08EE413 pKa = 4.67GGVGLAAYY421 pKa = 7.47TFADD425 pKa = 3.79ADD427 pKa = 3.71QSAMLAFFEE436 pKa = 5.58ALDD439 pKa = 3.81NDD441 pKa = 3.61PRR443 pKa = 11.84MGDD446 pKa = 3.23MYY448 pKa = 11.65AKK450 pKa = 9.72MVADD454 pKa = 5.34FAASGGSGLNAFTDD468 pKa = 3.59AGSPSKK474 pKa = 10.22WGTWGTLDD482 pKa = 4.26SIYY485 pKa = 10.91DD486 pKa = 3.89EE487 pKa = 4.72GSEE490 pKa = 4.73RR491 pKa = 11.84YY492 pKa = 9.03DD493 pKa = 3.48ALVAASQAAKK503 pKa = 10.12AAQLTGGDD511 pKa = 3.45ATQVNVLTSAASYY524 pKa = 11.08VLGGNQLTLTYY535 pKa = 10.3VGSGGFNGTGNDD547 pKa = 3.59LSNTIVGGNSFNMLYY562 pKa = 10.76GGGGDD567 pKa = 3.64DD568 pKa = 4.14TIVGGSRR575 pKa = 11.84NDD577 pKa = 3.64IIDD580 pKa = 3.86GGTGADD586 pKa = 3.53RR587 pKa = 11.84MIGGDD592 pKa = 3.58GDD594 pKa = 3.69DD595 pKa = 4.13TYY597 pKa = 12.03VVDD600 pKa = 4.54NIGDD604 pKa = 3.92VVVEE608 pKa = 4.0QAGGGTDD615 pKa = 3.52EE616 pKa = 5.5VRR618 pKa = 11.84TTLNTYY624 pKa = 10.92ALTASVEE631 pKa = 4.01NLTFIGTGVITGTGNDD647 pKa = 3.52SDD649 pKa = 4.84NIIAAVAGASASLYY663 pKa = 10.76GGAGNDD669 pKa = 3.57TLTGSTGADD678 pKa = 3.41LLDD681 pKa = 4.49GGTGADD687 pKa = 2.96IMRR690 pKa = 11.84GGRR693 pKa = 11.84GNDD696 pKa = 3.25VYY698 pKa = 11.04IVDD701 pKa = 3.77NAGDD705 pKa = 3.96NVIEE709 pKa = 4.12LAGGGIDD716 pKa = 4.05TVRR719 pKa = 11.84TTLNLYY725 pKa = 10.0GLASEE730 pKa = 4.7VEE732 pKa = 4.17NLTFIGTGNFNAFGNSLDD750 pKa = 3.7NVMTGGAGDD759 pKa = 3.62DD760 pKa = 4.13TLWGDD765 pKa = 4.23VGNDD769 pKa = 3.65TLNGGAGNDD778 pKa = 3.59VLIGGVGADD787 pKa = 3.63TMIGGLGDD795 pKa = 3.99DD796 pKa = 4.97RR797 pKa = 11.84YY798 pKa = 11.31DD799 pKa = 3.23VDD801 pKa = 4.27NIGDD805 pKa = 3.9IVVEE809 pKa = 4.76LPGQGNDD816 pKa = 3.42AVYY819 pKa = 8.13TTVNYY824 pKa = 9.19TLTANVEE831 pKa = 4.06NLMLSGTGNINGTGNEE847 pKa = 4.52LNNIMFGNSGDD858 pKa = 4.18NILSGGAGDD867 pKa = 4.03DD868 pKa = 3.83TLIGGAGNDD877 pKa = 3.8TLVGGDD883 pKa = 4.41GNDD886 pKa = 3.66SLDD889 pKa = 4.17GGDD892 pKa = 5.46GDD894 pKa = 6.07DD895 pKa = 4.07ILNGRR900 pKa = 11.84MGDD903 pKa = 3.76DD904 pKa = 5.63LITGGAGNDD913 pKa = 4.03VIWGGIGRR921 pKa = 11.84DD922 pKa = 3.76VMDD925 pKa = 5.1GGAGADD931 pKa = 3.16RR932 pKa = 11.84FIFYY936 pKa = 9.96TGDD939 pKa = 3.33TGKK942 pKa = 8.16TRR944 pKa = 11.84ATADD948 pKa = 3.36MIYY951 pKa = 10.31FSHH954 pKa = 7.4SDD956 pKa = 3.12GDD958 pKa = 4.56LIDD961 pKa = 4.63LSRR964 pKa = 11.84MDD966 pKa = 4.18ARR968 pKa = 11.84TATSVDD974 pKa = 3.51DD975 pKa = 4.07AFSFIGQAAFTNTAGEE991 pKa = 4.08LRR993 pKa = 11.84IQYY996 pKa = 10.35SGGYY1000 pKa = 8.54WDD1002 pKa = 5.23VLGDD1006 pKa = 3.76TNGDD1010 pKa = 3.88GIADD1014 pKa = 3.95FSITVSAGSTPLIARR1029 pKa = 11.84DD1030 pKa = 3.76FVLL1033 pKa = 5.2
MM1 pKa = 7.2NLAGVSYY8 pKa = 11.02YY9 pKa = 9.06GTQYY13 pKa = 11.15PFLDD17 pKa = 3.38RR18 pKa = 11.84FKK20 pKa = 10.64TSSEE24 pKa = 3.56WMVQGVNGQPIGALSLDD41 pKa = 3.48ANGYY45 pKa = 5.06PTAMPTGATNIFTMVGLDD63 pKa = 3.6PVALEE68 pKa = 4.02TSDD71 pKa = 4.1IYY73 pKa = 11.53VITYY77 pKa = 9.47KK78 pKa = 10.99GNASINILGGTILSSEE94 pKa = 4.15PGKK97 pKa = 9.24ITFRR101 pKa = 11.84FTGEE105 pKa = 4.03GSSTAINVSAIDD117 pKa = 3.73PANPLSDD124 pKa = 3.36MHH126 pKa = 6.97VVRR129 pKa = 11.84QDD131 pKa = 3.17QVALYY136 pKa = 10.03EE137 pKa = 4.33SGEE140 pKa = 4.07IFNPAFLDD148 pKa = 4.24KK149 pKa = 10.82ASQWSEE155 pKa = 3.69LRR157 pKa = 11.84YY158 pKa = 9.41MDD160 pKa = 4.85WGNTNNNTLTSWDD173 pKa = 3.42QRR175 pKa = 11.84TTLTSASWSNISSTHH190 pKa = 5.03SVPIEE195 pKa = 3.92IMVALANKK203 pKa = 8.67TNTDD207 pKa = 2.64MWINIPAAADD217 pKa = 3.23DD218 pKa = 4.23DD219 pKa = 4.98YY220 pKa = 11.7IRR222 pKa = 11.84KK223 pKa = 6.93TLSYY227 pKa = 11.14VRR229 pKa = 11.84DD230 pKa = 3.56NLAAGLSVKK239 pKa = 10.15VEE241 pKa = 4.02YY242 pKa = 11.14SNEE245 pKa = 3.31VWNWGFQQASYY256 pKa = 9.37AQTMGAQMFGTDD268 pKa = 4.09LNGNGQIDD276 pKa = 3.74RR277 pKa = 11.84GTSEE281 pKa = 4.43EE282 pKa = 4.57PGTAYY287 pKa = 10.33LQYY290 pKa = 9.98YY291 pKa = 8.72GYY293 pKa = 10.67RR294 pKa = 11.84AAQVAAVANSVFASSDD310 pKa = 3.29PARR313 pKa = 11.84LQNVLSTQTGYY324 pKa = 11.29LGLEE328 pKa = 4.02TYY330 pKa = 10.45IFDD333 pKa = 4.43GVQSAALGSVASLFDD348 pKa = 3.41NYY350 pKa = 11.28AITTYY355 pKa = 10.67FGLDD359 pKa = 3.23SVDD362 pKa = 3.39QATLVSWAHH371 pKa = 6.55AGQAGIDD378 pKa = 3.45AAFAGLNASLNDD390 pKa = 3.5SLRR393 pKa = 11.84VYY395 pKa = 10.5VYY397 pKa = 9.57QQAVAARR404 pKa = 11.84NGLSLVAYY412 pKa = 9.08EE413 pKa = 4.67GGVGLAAYY421 pKa = 7.47TFADD425 pKa = 3.79ADD427 pKa = 3.71QSAMLAFFEE436 pKa = 5.58ALDD439 pKa = 3.81NDD441 pKa = 3.61PRR443 pKa = 11.84MGDD446 pKa = 3.23MYY448 pKa = 11.65AKK450 pKa = 9.72MVADD454 pKa = 5.34FAASGGSGLNAFTDD468 pKa = 3.59AGSPSKK474 pKa = 10.22WGTWGTLDD482 pKa = 4.26SIYY485 pKa = 10.91DD486 pKa = 3.89EE487 pKa = 4.72GSEE490 pKa = 4.73RR491 pKa = 11.84YY492 pKa = 9.03DD493 pKa = 3.48ALVAASQAAKK503 pKa = 10.12AAQLTGGDD511 pKa = 3.45ATQVNVLTSAASYY524 pKa = 11.08VLGGNQLTLTYY535 pKa = 10.3VGSGGFNGTGNDD547 pKa = 3.59LSNTIVGGNSFNMLYY562 pKa = 10.76GGGGDD567 pKa = 3.64DD568 pKa = 4.14TIVGGSRR575 pKa = 11.84NDD577 pKa = 3.64IIDD580 pKa = 3.86GGTGADD586 pKa = 3.53RR587 pKa = 11.84MIGGDD592 pKa = 3.58GDD594 pKa = 3.69DD595 pKa = 4.13TYY597 pKa = 12.03VVDD600 pKa = 4.54NIGDD604 pKa = 3.92VVVEE608 pKa = 4.0QAGGGTDD615 pKa = 3.52EE616 pKa = 5.5VRR618 pKa = 11.84TTLNTYY624 pKa = 10.92ALTASVEE631 pKa = 4.01NLTFIGTGVITGTGNDD647 pKa = 3.52SDD649 pKa = 4.84NIIAAVAGASASLYY663 pKa = 10.76GGAGNDD669 pKa = 3.57TLTGSTGADD678 pKa = 3.41LLDD681 pKa = 4.49GGTGADD687 pKa = 2.96IMRR690 pKa = 11.84GGRR693 pKa = 11.84GNDD696 pKa = 3.25VYY698 pKa = 11.04IVDD701 pKa = 3.77NAGDD705 pKa = 3.96NVIEE709 pKa = 4.12LAGGGIDD716 pKa = 4.05TVRR719 pKa = 11.84TTLNLYY725 pKa = 10.0GLASEE730 pKa = 4.7VEE732 pKa = 4.17NLTFIGTGNFNAFGNSLDD750 pKa = 3.7NVMTGGAGDD759 pKa = 3.62DD760 pKa = 4.13TLWGDD765 pKa = 4.23VGNDD769 pKa = 3.65TLNGGAGNDD778 pKa = 3.59VLIGGVGADD787 pKa = 3.63TMIGGLGDD795 pKa = 3.99DD796 pKa = 4.97RR797 pKa = 11.84YY798 pKa = 11.31DD799 pKa = 3.23VDD801 pKa = 4.27NIGDD805 pKa = 3.9IVVEE809 pKa = 4.76LPGQGNDD816 pKa = 3.42AVYY819 pKa = 8.13TTVNYY824 pKa = 9.19TLTANVEE831 pKa = 4.06NLMLSGTGNINGTGNEE847 pKa = 4.52LNNIMFGNSGDD858 pKa = 4.18NILSGGAGDD867 pKa = 4.03DD868 pKa = 3.83TLIGGAGNDD877 pKa = 3.8TLVGGDD883 pKa = 4.41GNDD886 pKa = 3.66SLDD889 pKa = 4.17GGDD892 pKa = 5.46GDD894 pKa = 6.07DD895 pKa = 4.07ILNGRR900 pKa = 11.84MGDD903 pKa = 3.76DD904 pKa = 5.63LITGGAGNDD913 pKa = 4.03VIWGGIGRR921 pKa = 11.84DD922 pKa = 3.76VMDD925 pKa = 5.1GGAGADD931 pKa = 3.16RR932 pKa = 11.84FIFYY936 pKa = 9.96TGDD939 pKa = 3.33TGKK942 pKa = 8.16TRR944 pKa = 11.84ATADD948 pKa = 3.36MIYY951 pKa = 10.31FSHH954 pKa = 7.4SDD956 pKa = 3.12GDD958 pKa = 4.56LIDD961 pKa = 4.63LSRR964 pKa = 11.84MDD966 pKa = 4.18ARR968 pKa = 11.84TATSVDD974 pKa = 3.51DD975 pKa = 4.07AFSFIGQAAFTNTAGEE991 pKa = 4.08LRR993 pKa = 11.84IQYY996 pKa = 10.35SGGYY1000 pKa = 8.54WDD1002 pKa = 5.23VLGDD1006 pKa = 3.76TNGDD1010 pKa = 3.88GIADD1014 pKa = 3.95FSITVSAGSTPLIARR1029 pKa = 11.84DD1030 pKa = 3.76FVLL1033 pKa = 5.2
Molecular weight: 106.97 kDa
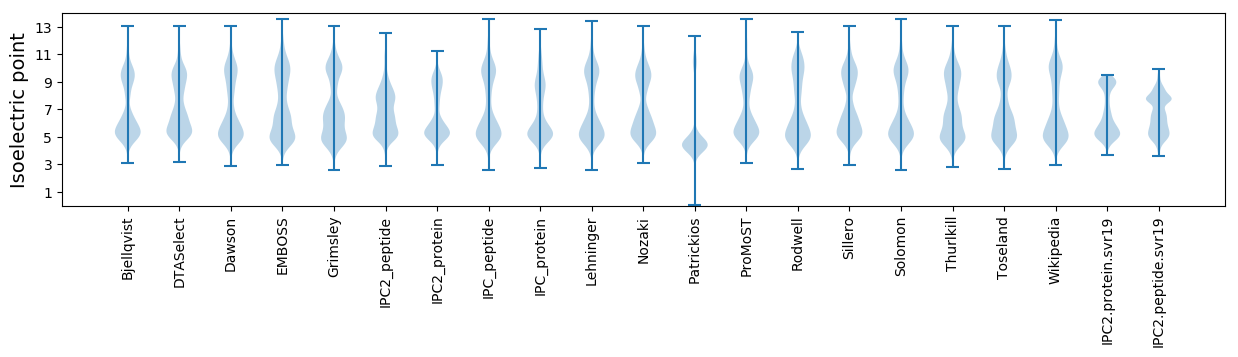
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q5QVU5|A0A0Q5QVU5_9SPHN Calpastatin OS=Sphingomonas sp. Leaf357 OX=1736350 GN=ASG11_04265 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.36GFRR19 pKa = 11.84ARR21 pKa = 11.84SATPGGRR28 pKa = 11.84NVLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.0KK41 pKa = 10.58LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.36GFRR19 pKa = 11.84ARR21 pKa = 11.84SATPGGRR28 pKa = 11.84NVLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.0KK41 pKa = 10.58LSAA44 pKa = 4.03
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1120184 |
29 |
1936 |
317.9 |
34.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.7 ± 0.069 | 0.728 ± 0.012 |
6.094 ± 0.03 | 4.883 ± 0.037 |
3.545 ± 0.028 | 9.072 ± 0.035 |
1.899 ± 0.02 | 5.162 ± 0.026 |
3.041 ± 0.031 | 9.689 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.401 ± 0.021 | 2.531 ± 0.031 |
5.32 ± 0.029 | 2.933 ± 0.022 |
7.173 ± 0.04 | 5.093 ± 0.025 |
5.687 ± 0.036 | 7.388 ± 0.033 |
1.405 ± 0.019 | 2.257 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
