
Enterobacteria phage M (Escherichia levivirus M)
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Leviviricetes; Norzivirales; Fiersviridae; Empivirus; Empivirus allolyticum
Average proteome isoelectric point is 8.55
Get precalculated fractions of proteins
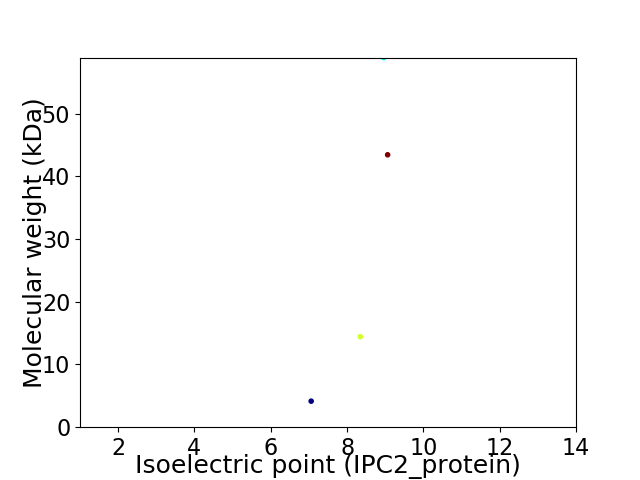
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
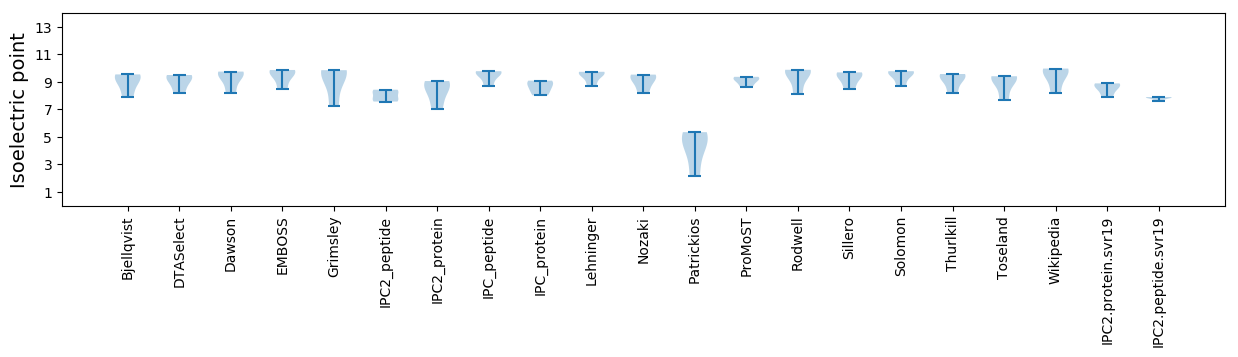
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7QKZ0|K7QKZ0_BPM RNA replicase beta chain OS=Enterobacteria phage M OX=1235640 GN=rep PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 10.56YY3 pKa = 10.13IINLAFCVLLLVAGDD18 pKa = 4.26SIAYY22 pKa = 9.29RR23 pKa = 11.84VSQYY27 pKa = 10.73LAPLVDD33 pKa = 3.99TFTKK37 pKa = 10.7
MM1 pKa = 7.66KK2 pKa = 10.56YY3 pKa = 10.13IINLAFCVLLLVAGDD18 pKa = 4.26SIAYY22 pKa = 9.29RR23 pKa = 11.84VSQYY27 pKa = 10.73LAPLVDD33 pKa = 3.99TFTKK37 pKa = 10.7
Molecular weight: 4.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7QLN7|K7QLN7_BPM Maturation protein OS=Enterobacteria phage M OX=1235640 GN=mat PE=4 SV=1
MM1 pKa = 6.87SRR3 pKa = 11.84KK4 pKa = 9.38RR5 pKa = 11.84RR6 pKa = 11.84VTISTSQFHH15 pKa = 5.88VQLDD19 pKa = 4.18EE20 pKa = 3.91QWVLRR25 pKa = 11.84RR26 pKa = 11.84ISEE29 pKa = 4.24SLDD32 pKa = 3.15LSDD35 pKa = 5.29FRR37 pKa = 11.84NSYY40 pKa = 9.6LVKK43 pKa = 10.67SLLSKK48 pKa = 10.48YY49 pKa = 9.6AAPSKK54 pKa = 10.04EE55 pKa = 4.09SARR58 pKa = 11.84ARR60 pKa = 11.84QRR62 pKa = 11.84AAIAKK67 pKa = 8.09LLWTSRR73 pKa = 11.84RR74 pKa = 11.84NRR76 pKa = 11.84DD77 pKa = 2.75TEE79 pKa = 3.88YY80 pKa = 11.05RR81 pKa = 11.84LLRR84 pKa = 11.84RR85 pKa = 11.84DD86 pKa = 3.41SKK88 pKa = 11.23QDD90 pKa = 3.18FMFLLTRR97 pKa = 11.84EE98 pKa = 4.26VANLLGDD105 pKa = 3.62VPEE108 pKa = 4.5FEE110 pKa = 4.84GLSSFSGGASTRR122 pKa = 11.84HH123 pKa = 5.48RR124 pKa = 11.84RR125 pKa = 11.84GKK127 pKa = 9.58SQPQEE132 pKa = 4.04KK133 pKa = 10.02FDD135 pKa = 4.31GYY137 pKa = 11.73GDD139 pKa = 3.71TTASNLPLARR149 pKa = 11.84KK150 pKa = 9.85SILASPAWSCNSAVSTGGTLEE171 pKa = 4.07LRR173 pKa = 11.84LVPGNVVFTVPKK185 pKa = 10.54SNVIDD190 pKa = 3.47RR191 pKa = 11.84AAAKK195 pKa = 10.45EE196 pKa = 4.13PDD198 pKa = 3.31LNMYY202 pKa = 7.27VQKK205 pKa = 10.79FYY207 pKa = 11.33GDD209 pKa = 4.32HH210 pKa = 5.19IRR212 pKa = 11.84GRR214 pKa = 11.84LRR216 pKa = 11.84RR217 pKa = 11.84FGIDD221 pKa = 3.91LNDD224 pKa = 3.33QGRR227 pKa = 11.84NRR229 pKa = 11.84DD230 pKa = 3.47LARR233 pKa = 11.84QGSITGDD240 pKa = 3.6LATLDD245 pKa = 4.36LSSASDD251 pKa = 3.8SITRR255 pKa = 11.84ILMLEE260 pKa = 4.13VLPPAWFDD268 pKa = 3.35VLDD271 pKa = 3.97RR272 pKa = 11.84ARR274 pKa = 11.84SRR276 pKa = 11.84WTNVNGVYY284 pKa = 9.96RR285 pKa = 11.84QLHH288 pKa = 5.45MFSTMGNGFTFEE300 pKa = 4.6LEE302 pKa = 4.07SLLFFAVTRR311 pKa = 11.84VTLKK315 pKa = 9.86MLNIQGTIGIYY326 pKa = 10.53GDD328 pKa = 5.78DD329 pKa = 4.27IICPTSAARR338 pKa = 11.84EE339 pKa = 4.14VVSNLEE345 pKa = 4.08YY346 pKa = 10.49IGFKK350 pKa = 10.52TNIDD354 pKa = 3.1KK355 pKa = 11.34SFIEE359 pKa = 3.96GHH361 pKa = 5.42FRR363 pKa = 11.84EE364 pKa = 4.79SCGGHH369 pKa = 5.43YY370 pKa = 10.62YY371 pKa = 10.52KK372 pKa = 11.01GRR374 pKa = 11.84DD375 pKa = 3.23VTPFYY380 pKa = 10.68IRR382 pKa = 11.84SPLNNIQRR390 pKa = 11.84IIWLLNKK397 pKa = 9.28IRR399 pKa = 11.84EE400 pKa = 4.23WSSEE404 pKa = 3.64GTTIAVEE411 pKa = 3.98LEE413 pKa = 3.73EE414 pKa = 5.71LYY416 pKa = 11.12FEE418 pKa = 5.11IYY420 pKa = 10.77NKK422 pKa = 10.2FSILRR427 pKa = 11.84SLTGCGGLDD436 pKa = 3.84SISSLAIPGAFGGYY450 pKa = 8.35LHH452 pKa = 6.46QVTRR456 pKa = 11.84RR457 pKa = 11.84VAVNEE462 pKa = 4.04DD463 pKa = 3.08GAYY466 pKa = 9.92VQALLKK472 pKa = 10.6GSPSDD477 pKa = 3.24TLRR480 pKa = 11.84FVSVEE485 pKa = 3.59TSRR488 pKa = 11.84YY489 pKa = 9.44EE490 pKa = 3.5IRR492 pKa = 11.84QWSDD496 pKa = 2.32KK497 pKa = 10.28VRR499 pKa = 11.84PDD501 pKa = 3.22MCRR504 pKa = 11.84SLYY507 pKa = 10.78SNGRR511 pKa = 11.84MVYY514 pKa = 9.6FLRR517 pKa = 11.84EE518 pKa = 3.73IGG520 pKa = 3.46
MM1 pKa = 6.87SRR3 pKa = 11.84KK4 pKa = 9.38RR5 pKa = 11.84RR6 pKa = 11.84VTISTSQFHH15 pKa = 5.88VQLDD19 pKa = 4.18EE20 pKa = 3.91QWVLRR25 pKa = 11.84RR26 pKa = 11.84ISEE29 pKa = 4.24SLDD32 pKa = 3.15LSDD35 pKa = 5.29FRR37 pKa = 11.84NSYY40 pKa = 9.6LVKK43 pKa = 10.67SLLSKK48 pKa = 10.48YY49 pKa = 9.6AAPSKK54 pKa = 10.04EE55 pKa = 4.09SARR58 pKa = 11.84ARR60 pKa = 11.84QRR62 pKa = 11.84AAIAKK67 pKa = 8.09LLWTSRR73 pKa = 11.84RR74 pKa = 11.84NRR76 pKa = 11.84DD77 pKa = 2.75TEE79 pKa = 3.88YY80 pKa = 11.05RR81 pKa = 11.84LLRR84 pKa = 11.84RR85 pKa = 11.84DD86 pKa = 3.41SKK88 pKa = 11.23QDD90 pKa = 3.18FMFLLTRR97 pKa = 11.84EE98 pKa = 4.26VANLLGDD105 pKa = 3.62VPEE108 pKa = 4.5FEE110 pKa = 4.84GLSSFSGGASTRR122 pKa = 11.84HH123 pKa = 5.48RR124 pKa = 11.84RR125 pKa = 11.84GKK127 pKa = 9.58SQPQEE132 pKa = 4.04KK133 pKa = 10.02FDD135 pKa = 4.31GYY137 pKa = 11.73GDD139 pKa = 3.71TTASNLPLARR149 pKa = 11.84KK150 pKa = 9.85SILASPAWSCNSAVSTGGTLEE171 pKa = 4.07LRR173 pKa = 11.84LVPGNVVFTVPKK185 pKa = 10.54SNVIDD190 pKa = 3.47RR191 pKa = 11.84AAAKK195 pKa = 10.45EE196 pKa = 4.13PDD198 pKa = 3.31LNMYY202 pKa = 7.27VQKK205 pKa = 10.79FYY207 pKa = 11.33GDD209 pKa = 4.32HH210 pKa = 5.19IRR212 pKa = 11.84GRR214 pKa = 11.84LRR216 pKa = 11.84RR217 pKa = 11.84FGIDD221 pKa = 3.91LNDD224 pKa = 3.33QGRR227 pKa = 11.84NRR229 pKa = 11.84DD230 pKa = 3.47LARR233 pKa = 11.84QGSITGDD240 pKa = 3.6LATLDD245 pKa = 4.36LSSASDD251 pKa = 3.8SITRR255 pKa = 11.84ILMLEE260 pKa = 4.13VLPPAWFDD268 pKa = 3.35VLDD271 pKa = 3.97RR272 pKa = 11.84ARR274 pKa = 11.84SRR276 pKa = 11.84WTNVNGVYY284 pKa = 9.96RR285 pKa = 11.84QLHH288 pKa = 5.45MFSTMGNGFTFEE300 pKa = 4.6LEE302 pKa = 4.07SLLFFAVTRR311 pKa = 11.84VTLKK315 pKa = 9.86MLNIQGTIGIYY326 pKa = 10.53GDD328 pKa = 5.78DD329 pKa = 4.27IICPTSAARR338 pKa = 11.84EE339 pKa = 4.14VVSNLEE345 pKa = 4.08YY346 pKa = 10.49IGFKK350 pKa = 10.52TNIDD354 pKa = 3.1KK355 pKa = 11.34SFIEE359 pKa = 3.96GHH361 pKa = 5.42FRR363 pKa = 11.84EE364 pKa = 4.79SCGGHH369 pKa = 5.43YY370 pKa = 10.62YY371 pKa = 10.52KK372 pKa = 11.01GRR374 pKa = 11.84DD375 pKa = 3.23VTPFYY380 pKa = 10.68IRR382 pKa = 11.84SPLNNIQRR390 pKa = 11.84IIWLLNKK397 pKa = 9.28IRR399 pKa = 11.84EE400 pKa = 4.23WSSEE404 pKa = 3.64GTTIAVEE411 pKa = 3.98LEE413 pKa = 3.73EE414 pKa = 5.71LYY416 pKa = 11.12FEE418 pKa = 5.11IYY420 pKa = 10.77NKK422 pKa = 10.2FSILRR427 pKa = 11.84SLTGCGGLDD436 pKa = 3.84SISSLAIPGAFGGYY450 pKa = 8.35LHH452 pKa = 6.46QVTRR456 pKa = 11.84RR457 pKa = 11.84VAVNEE462 pKa = 4.04DD463 pKa = 3.08GAYY466 pKa = 9.92VQALLKK472 pKa = 10.6GSPSDD477 pKa = 3.24TLRR480 pKa = 11.84FVSVEE485 pKa = 3.59TSRR488 pKa = 11.84YY489 pKa = 9.44EE490 pKa = 3.5IRR492 pKa = 11.84QWSDD496 pKa = 2.32KK497 pKa = 10.28VRR499 pKa = 11.84PDD501 pKa = 3.22MCRR504 pKa = 11.84SLYY507 pKa = 10.78SNGRR511 pKa = 11.84MVYY514 pKa = 9.6FLRR517 pKa = 11.84EE518 pKa = 3.73IGG520 pKa = 3.46
Molecular weight: 58.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1085 |
37 |
520 |
271.3 |
30.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.912 ± 0.722 | 1.014 ± 0.216 |
4.885 ± 0.525 | 4.24 ± 0.767 |
4.7 ± 0.167 | 7.373 ± 0.639 |
1.382 ± 0.443 | 5.53 ± 0.32 |
4.055 ± 0.522 | 9.862 ± 0.742 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.843 ± 0.164 | 4.24 ± 0.259 |
4.424 ± 0.868 | 2.673 ± 0.599 |
7.926 ± 1.307 | 10.138 ± 0.903 |
6.82 ± 0.753 | 6.636 ± 1.467 |
1.475 ± 0.448 | 3.871 ± 0.458 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
