
Clostridium sp. CAG:628
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; environmental samples
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
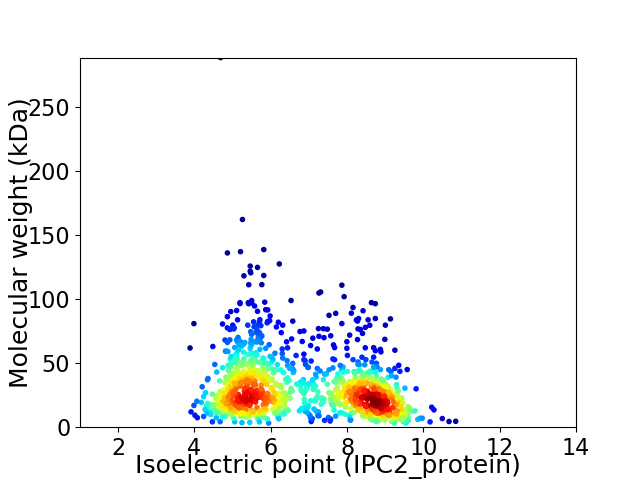
Virtual 2D-PAGE plot for 1091 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7MFX6|R7MFX6_9CLOT Uncharacterized protein OS=Clostridium sp. CAG:628 OX=1262829 GN=BN740_01045 PE=4 SV=1
MM1 pKa = 7.56NDD3 pKa = 3.54DD4 pKa = 3.5EE5 pKa = 5.08KK6 pKa = 11.38KK7 pKa = 10.41IIEE10 pKa = 4.14EE11 pKa = 3.75EE12 pKa = 3.79GLYY15 pKa = 10.31FVEE18 pKa = 5.8PEE20 pKa = 4.02DD21 pKa = 3.71KK22 pKa = 10.4KK23 pKa = 10.82KK24 pKa = 10.67KK25 pKa = 9.7RR26 pKa = 11.84VIIIILLLILLLIAIGSASFSVYY49 pKa = 10.87NYY51 pKa = 9.72FKK53 pKa = 10.4IYY55 pKa = 10.22KK56 pKa = 9.3HH57 pKa = 6.33RR58 pKa = 11.84LNIDD62 pKa = 3.46TDD64 pKa = 3.26WDD66 pKa = 4.52GIADD70 pKa = 4.56LNVDD74 pKa = 4.48LDD76 pKa = 4.0NDD78 pKa = 4.21GKK80 pKa = 11.32CDD82 pKa = 3.88VNCDD86 pKa = 3.15TSGNGKK92 pKa = 9.01PDD94 pKa = 3.72LNVGYY99 pKa = 10.48KK100 pKa = 9.96KK101 pKa = 10.56VLKK104 pKa = 10.08PYY106 pKa = 10.61FNIDD110 pKa = 2.9TDD112 pKa = 4.48GDD114 pKa = 4.0GKK116 pKa = 10.02PDD118 pKa = 3.38KK119 pKa = 11.07NLINVKK125 pKa = 10.66DD126 pKa = 3.96EE127 pKa = 4.52DD128 pKa = 4.04GKK130 pKa = 11.24CIKK133 pKa = 10.86NCDD136 pKa = 4.03TNNDD140 pKa = 3.98GYY142 pKa = 10.06PDD144 pKa = 3.84TNIDD148 pKa = 3.68FDD150 pKa = 5.9DD151 pKa = 6.34DD152 pKa = 5.06GICDD156 pKa = 4.74LNCDD160 pKa = 3.31TDD162 pKa = 4.79KK163 pKa = 11.57DD164 pKa = 4.29GKK166 pKa = 10.95CDD168 pKa = 3.65VNCDD172 pKa = 3.34VNGDD176 pKa = 3.99GKK178 pKa = 11.23CDD180 pKa = 4.1LNCDD184 pKa = 3.4TDD186 pKa = 4.8KK187 pKa = 11.48DD188 pKa = 4.22GKK190 pKa = 10.0PDD192 pKa = 3.59TNLDD196 pKa = 3.43EE197 pKa = 5.82DD198 pKa = 4.84KK199 pKa = 11.52DD200 pKa = 4.46GICDD204 pKa = 3.67KK205 pKa = 11.32NCTGKK210 pKa = 10.78EE211 pKa = 3.7NDD213 pKa = 3.43VCKK216 pKa = 10.64INCDD220 pKa = 3.4TNGDD224 pKa = 3.94GKK226 pKa = 10.8CDD228 pKa = 4.35RR229 pKa = 11.84NCDD232 pKa = 3.83DD233 pKa = 5.23NKK235 pKa = 11.36DD236 pKa = 3.88GVCDD240 pKa = 4.07RR241 pKa = 11.84NCDD244 pKa = 3.3VDD246 pKa = 3.81GDD248 pKa = 4.84GYY250 pKa = 11.58CEE252 pKa = 4.53LNCDD256 pKa = 3.33TKK258 pKa = 11.74GNGKK262 pKa = 9.48CDD264 pKa = 3.87KK265 pKa = 11.32NCDD268 pKa = 3.4TDD270 pKa = 4.43NDD272 pKa = 5.1GICNVKK278 pKa = 10.32CGNTGKK284 pKa = 10.21DD285 pKa = 3.29LCLVNCDD292 pKa = 3.13TDD294 pKa = 5.02GDD296 pKa = 4.41GKK298 pKa = 10.83CDD300 pKa = 4.28KK301 pKa = 11.1NCDD304 pKa = 3.57YY305 pKa = 11.49NGDD308 pKa = 3.9GKK310 pKa = 11.24CDD312 pKa = 4.17LNCDD316 pKa = 3.55TNNNGKK322 pKa = 9.53CDD324 pKa = 4.0KK325 pKa = 11.35NCDD328 pKa = 3.67TNNDD332 pKa = 4.33GICDD336 pKa = 3.92KK337 pKa = 11.23KK338 pKa = 11.18CDD340 pKa = 3.78GNEE343 pKa = 3.87NDD345 pKa = 4.14SCKK348 pKa = 10.65LNCDD352 pKa = 3.57FNGDD356 pKa = 3.81GKK358 pKa = 10.9CDD360 pKa = 3.88KK361 pKa = 11.33NCDD364 pKa = 3.2TDD366 pKa = 5.36GDD368 pKa = 4.39GKK370 pKa = 10.78CDD372 pKa = 3.88KK373 pKa = 11.04NCDD376 pKa = 3.45VNGDD380 pKa = 3.83GKK382 pKa = 10.95CDD384 pKa = 3.77YY385 pKa = 11.4NCTEE389 pKa = 4.36NNDD392 pKa = 3.56VCLKK396 pKa = 10.77NCDD399 pKa = 3.76LNGDD403 pKa = 4.37GKK405 pKa = 11.25CDD407 pKa = 4.46LNCDD411 pKa = 3.3TDD413 pKa = 4.67NDD415 pKa = 4.31GKK417 pKa = 10.88CDD419 pKa = 4.04KK420 pKa = 11.41NCDD423 pKa = 3.4TDD425 pKa = 4.37NDD427 pKa = 5.23GICDD431 pKa = 3.83KK432 pKa = 11.48NCDD435 pKa = 3.39GKK437 pKa = 11.46EE438 pKa = 3.86NDD440 pKa = 3.48VCKK443 pKa = 10.64INCDD447 pKa = 3.4TNGDD451 pKa = 3.94GKK453 pKa = 10.79CDD455 pKa = 3.91RR456 pKa = 11.84NCDD459 pKa = 3.17TDD461 pKa = 4.92NDD463 pKa = 4.37GKK465 pKa = 10.94CDD467 pKa = 3.91KK468 pKa = 11.22NCDD471 pKa = 3.45ANGDD475 pKa = 3.88GKK477 pKa = 11.02CDD479 pKa = 3.81YY480 pKa = 11.06NCDD483 pKa = 3.83AGSDD487 pKa = 3.48KK488 pKa = 10.98CKK490 pKa = 10.51RR491 pKa = 11.84NCDD494 pKa = 3.21ADD496 pKa = 4.13NDD498 pKa = 4.53GKK500 pKa = 11.33CEE502 pKa = 4.47LNCDD506 pKa = 3.78TNNDD510 pKa = 3.66GVCDD514 pKa = 4.39KK515 pKa = 11.53NCDD518 pKa = 3.36TDD520 pKa = 4.71GDD522 pKa = 4.67GICDD526 pKa = 3.73KK527 pKa = 11.45NCDD530 pKa = 3.36GKK532 pKa = 11.49EE533 pKa = 3.83NDD535 pKa = 3.64TCKK538 pKa = 10.48INCDD542 pKa = 3.11TDD544 pKa = 4.13NDD546 pKa = 4.32GKK548 pKa = 10.88CDD550 pKa = 4.31KK551 pKa = 11.41NCDD554 pKa = 3.65TNNDD558 pKa = 3.62GVCDD562 pKa = 4.39KK563 pKa = 11.32NCDD566 pKa = 3.64SNGDD570 pKa = 3.75GKK572 pKa = 11.26CDD574 pKa = 4.12LNCDD578 pKa = 2.88IDD580 pKa = 5.59GDD582 pKa = 4.39GKK584 pKa = 10.88CDD586 pKa = 4.33KK587 pKa = 11.31NCDD590 pKa = 3.65TNGDD594 pKa = 4.35GICDD598 pKa = 4.05KK599 pKa = 11.29NCGGPVDD606 pKa = 3.3IGKK609 pKa = 9.54NDD611 pKa = 3.49EE612 pKa = 4.58YY613 pKa = 10.98ILTFKK618 pKa = 11.01DD619 pKa = 2.85ITDD622 pKa = 4.13LKK624 pKa = 10.78NVNIYY629 pKa = 9.88PGWSASKK636 pKa = 10.67VFEE639 pKa = 4.2ITNTSAVTLTYY650 pKa = 10.55NINFTSVKK658 pKa = 10.8NEE660 pKa = 3.44FSTTNNLYY668 pKa = 10.81YY669 pKa = 11.02GLIKK673 pKa = 10.57DD674 pKa = 4.1NKK676 pKa = 8.56TLIDD680 pKa = 3.8INTARR685 pKa = 11.84APYY688 pKa = 9.89KK689 pKa = 10.57DD690 pKa = 3.24GVMLKK695 pKa = 10.51SIIIKK700 pKa = 10.04PGEE703 pKa = 3.78THH705 pKa = 6.84TYY707 pKa = 8.32MFKK710 pKa = 11.11LMFKK714 pKa = 10.66DD715 pKa = 3.13SDD717 pKa = 3.83VNQDD721 pKa = 2.47IDD723 pKa = 3.53KK724 pKa = 11.24GKK726 pKa = 10.18NFYY729 pKa = 10.42TKK731 pKa = 9.65IQIEE735 pKa = 4.35NVHH738 pKa = 6.02
MM1 pKa = 7.56NDD3 pKa = 3.54DD4 pKa = 3.5EE5 pKa = 5.08KK6 pKa = 11.38KK7 pKa = 10.41IIEE10 pKa = 4.14EE11 pKa = 3.75EE12 pKa = 3.79GLYY15 pKa = 10.31FVEE18 pKa = 5.8PEE20 pKa = 4.02DD21 pKa = 3.71KK22 pKa = 10.4KK23 pKa = 10.82KK24 pKa = 10.67KK25 pKa = 9.7RR26 pKa = 11.84VIIIILLLILLLIAIGSASFSVYY49 pKa = 10.87NYY51 pKa = 9.72FKK53 pKa = 10.4IYY55 pKa = 10.22KK56 pKa = 9.3HH57 pKa = 6.33RR58 pKa = 11.84LNIDD62 pKa = 3.46TDD64 pKa = 3.26WDD66 pKa = 4.52GIADD70 pKa = 4.56LNVDD74 pKa = 4.48LDD76 pKa = 4.0NDD78 pKa = 4.21GKK80 pKa = 11.32CDD82 pKa = 3.88VNCDD86 pKa = 3.15TSGNGKK92 pKa = 9.01PDD94 pKa = 3.72LNVGYY99 pKa = 10.48KK100 pKa = 9.96KK101 pKa = 10.56VLKK104 pKa = 10.08PYY106 pKa = 10.61FNIDD110 pKa = 2.9TDD112 pKa = 4.48GDD114 pKa = 4.0GKK116 pKa = 10.02PDD118 pKa = 3.38KK119 pKa = 11.07NLINVKK125 pKa = 10.66DD126 pKa = 3.96EE127 pKa = 4.52DD128 pKa = 4.04GKK130 pKa = 11.24CIKK133 pKa = 10.86NCDD136 pKa = 4.03TNNDD140 pKa = 3.98GYY142 pKa = 10.06PDD144 pKa = 3.84TNIDD148 pKa = 3.68FDD150 pKa = 5.9DD151 pKa = 6.34DD152 pKa = 5.06GICDD156 pKa = 4.74LNCDD160 pKa = 3.31TDD162 pKa = 4.79KK163 pKa = 11.57DD164 pKa = 4.29GKK166 pKa = 10.95CDD168 pKa = 3.65VNCDD172 pKa = 3.34VNGDD176 pKa = 3.99GKK178 pKa = 11.23CDD180 pKa = 4.1LNCDD184 pKa = 3.4TDD186 pKa = 4.8KK187 pKa = 11.48DD188 pKa = 4.22GKK190 pKa = 10.0PDD192 pKa = 3.59TNLDD196 pKa = 3.43EE197 pKa = 5.82DD198 pKa = 4.84KK199 pKa = 11.52DD200 pKa = 4.46GICDD204 pKa = 3.67KK205 pKa = 11.32NCTGKK210 pKa = 10.78EE211 pKa = 3.7NDD213 pKa = 3.43VCKK216 pKa = 10.64INCDD220 pKa = 3.4TNGDD224 pKa = 3.94GKK226 pKa = 10.8CDD228 pKa = 4.35RR229 pKa = 11.84NCDD232 pKa = 3.83DD233 pKa = 5.23NKK235 pKa = 11.36DD236 pKa = 3.88GVCDD240 pKa = 4.07RR241 pKa = 11.84NCDD244 pKa = 3.3VDD246 pKa = 3.81GDD248 pKa = 4.84GYY250 pKa = 11.58CEE252 pKa = 4.53LNCDD256 pKa = 3.33TKK258 pKa = 11.74GNGKK262 pKa = 9.48CDD264 pKa = 3.87KK265 pKa = 11.32NCDD268 pKa = 3.4TDD270 pKa = 4.43NDD272 pKa = 5.1GICNVKK278 pKa = 10.32CGNTGKK284 pKa = 10.21DD285 pKa = 3.29LCLVNCDD292 pKa = 3.13TDD294 pKa = 5.02GDD296 pKa = 4.41GKK298 pKa = 10.83CDD300 pKa = 4.28KK301 pKa = 11.1NCDD304 pKa = 3.57YY305 pKa = 11.49NGDD308 pKa = 3.9GKK310 pKa = 11.24CDD312 pKa = 4.17LNCDD316 pKa = 3.55TNNNGKK322 pKa = 9.53CDD324 pKa = 4.0KK325 pKa = 11.35NCDD328 pKa = 3.67TNNDD332 pKa = 4.33GICDD336 pKa = 3.92KK337 pKa = 11.23KK338 pKa = 11.18CDD340 pKa = 3.78GNEE343 pKa = 3.87NDD345 pKa = 4.14SCKK348 pKa = 10.65LNCDD352 pKa = 3.57FNGDD356 pKa = 3.81GKK358 pKa = 10.9CDD360 pKa = 3.88KK361 pKa = 11.33NCDD364 pKa = 3.2TDD366 pKa = 5.36GDD368 pKa = 4.39GKK370 pKa = 10.78CDD372 pKa = 3.88KK373 pKa = 11.04NCDD376 pKa = 3.45VNGDD380 pKa = 3.83GKK382 pKa = 10.95CDD384 pKa = 3.77YY385 pKa = 11.4NCTEE389 pKa = 4.36NNDD392 pKa = 3.56VCLKK396 pKa = 10.77NCDD399 pKa = 3.76LNGDD403 pKa = 4.37GKK405 pKa = 11.25CDD407 pKa = 4.46LNCDD411 pKa = 3.3TDD413 pKa = 4.67NDD415 pKa = 4.31GKK417 pKa = 10.88CDD419 pKa = 4.04KK420 pKa = 11.41NCDD423 pKa = 3.4TDD425 pKa = 4.37NDD427 pKa = 5.23GICDD431 pKa = 3.83KK432 pKa = 11.48NCDD435 pKa = 3.39GKK437 pKa = 11.46EE438 pKa = 3.86NDD440 pKa = 3.48VCKK443 pKa = 10.64INCDD447 pKa = 3.4TNGDD451 pKa = 3.94GKK453 pKa = 10.79CDD455 pKa = 3.91RR456 pKa = 11.84NCDD459 pKa = 3.17TDD461 pKa = 4.92NDD463 pKa = 4.37GKK465 pKa = 10.94CDD467 pKa = 3.91KK468 pKa = 11.22NCDD471 pKa = 3.45ANGDD475 pKa = 3.88GKK477 pKa = 11.02CDD479 pKa = 3.81YY480 pKa = 11.06NCDD483 pKa = 3.83AGSDD487 pKa = 3.48KK488 pKa = 10.98CKK490 pKa = 10.51RR491 pKa = 11.84NCDD494 pKa = 3.21ADD496 pKa = 4.13NDD498 pKa = 4.53GKK500 pKa = 11.33CEE502 pKa = 4.47LNCDD506 pKa = 3.78TNNDD510 pKa = 3.66GVCDD514 pKa = 4.39KK515 pKa = 11.53NCDD518 pKa = 3.36TDD520 pKa = 4.71GDD522 pKa = 4.67GICDD526 pKa = 3.73KK527 pKa = 11.45NCDD530 pKa = 3.36GKK532 pKa = 11.49EE533 pKa = 3.83NDD535 pKa = 3.64TCKK538 pKa = 10.48INCDD542 pKa = 3.11TDD544 pKa = 4.13NDD546 pKa = 4.32GKK548 pKa = 10.88CDD550 pKa = 4.31KK551 pKa = 11.41NCDD554 pKa = 3.65TNNDD558 pKa = 3.62GVCDD562 pKa = 4.39KK563 pKa = 11.32NCDD566 pKa = 3.64SNGDD570 pKa = 3.75GKK572 pKa = 11.26CDD574 pKa = 4.12LNCDD578 pKa = 2.88IDD580 pKa = 5.59GDD582 pKa = 4.39GKK584 pKa = 10.88CDD586 pKa = 4.33KK587 pKa = 11.31NCDD590 pKa = 3.65TNGDD594 pKa = 4.35GICDD598 pKa = 4.05KK599 pKa = 11.29NCGGPVDD606 pKa = 3.3IGKK609 pKa = 9.54NDD611 pKa = 3.49EE612 pKa = 4.58YY613 pKa = 10.98ILTFKK618 pKa = 11.01DD619 pKa = 2.85ITDD622 pKa = 4.13LKK624 pKa = 10.78NVNIYY629 pKa = 9.88PGWSASKK636 pKa = 10.67VFEE639 pKa = 4.2ITNTSAVTLTYY650 pKa = 10.55NINFTSVKK658 pKa = 10.8NEE660 pKa = 3.44FSTTNNLYY668 pKa = 10.81YY669 pKa = 11.02GLIKK673 pKa = 10.57DD674 pKa = 4.1NKK676 pKa = 8.56TLIDD680 pKa = 3.8INTARR685 pKa = 11.84APYY688 pKa = 9.89KK689 pKa = 10.57DD690 pKa = 3.24GVMLKK695 pKa = 10.51SIIIKK700 pKa = 10.04PGEE703 pKa = 3.78THH705 pKa = 6.84TYY707 pKa = 8.32MFKK710 pKa = 11.11LMFKK714 pKa = 10.66DD715 pKa = 3.13SDD717 pKa = 3.83VNQDD721 pKa = 2.47IDD723 pKa = 3.53KK724 pKa = 11.24GKK726 pKa = 10.18NFYY729 pKa = 10.42TKK731 pKa = 9.65IQIEE735 pKa = 4.35NVHH738 pKa = 6.02
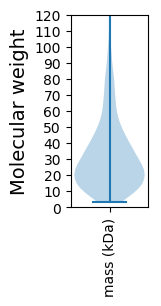
Molecular weight: 80.83 kDa
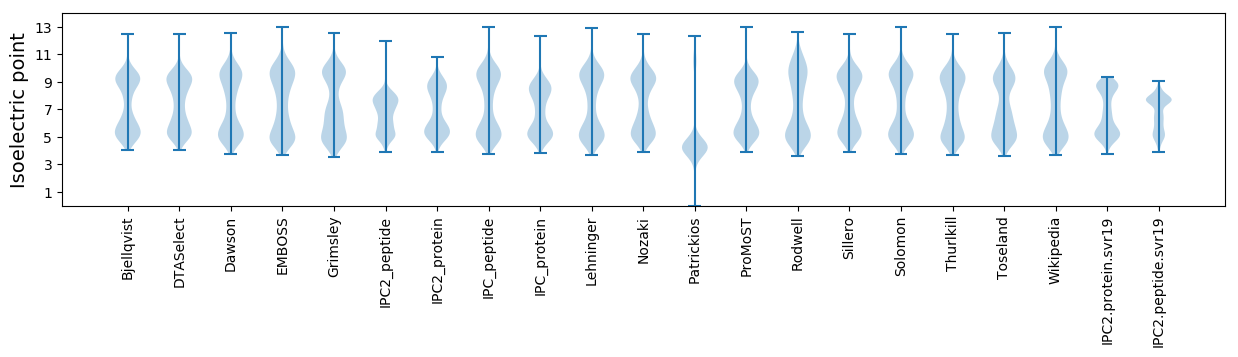
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7MIU9|R7MIU9_9CLOT Peptide chain release factor 1 OS=Clostridium sp. CAG:628 OX=1262829 GN=prfA PE=3 SV=1
MM1 pKa = 7.7KK2 pKa = 8.92LTPVAKK8 pKa = 10.37KK9 pKa = 10.23NNKK12 pKa = 9.04AKK14 pKa = 10.57KK15 pKa = 9.62QGFLSRR21 pKa = 11.84SANILKK27 pKa = 10.2SRR29 pKa = 11.84RR30 pKa = 11.84QKK32 pKa = 10.44GRR34 pKa = 11.84KK35 pKa = 8.53NLINKK40 pKa = 8.51
MM1 pKa = 7.7KK2 pKa = 8.92LTPVAKK8 pKa = 10.37KK9 pKa = 10.23NNKK12 pKa = 9.04AKK14 pKa = 10.57KK15 pKa = 9.62QGFLSRR21 pKa = 11.84SANILKK27 pKa = 10.2SRR29 pKa = 11.84RR30 pKa = 11.84QKK32 pKa = 10.44GRR34 pKa = 11.84KK35 pKa = 8.53NLINKK40 pKa = 8.51
Molecular weight: 4.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
319958 |
29 |
2658 |
293.3 |
33.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.575 ± 0.049 | 1.055 ± 0.036 |
5.767 ± 0.069 | 7.315 ± 0.09 |
4.281 ± 0.055 | 5.294 ± 0.087 |
1.141 ± 0.026 | 9.73 ± 0.082 |
10.114 ± 0.095 | 9.477 ± 0.092 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.389 ± 0.036 | 7.884 ± 0.093 |
2.323 ± 0.036 | 1.534 ± 0.029 |
2.967 ± 0.051 | 6.949 ± 0.077 |
5.762 ± 0.085 | 6.506 ± 0.072 |
0.512 ± 0.02 | 5.426 ± 0.066 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
