
Sus scrofa papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyodeltapapillomavirus; Dyodeltapapillomavirus 1
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
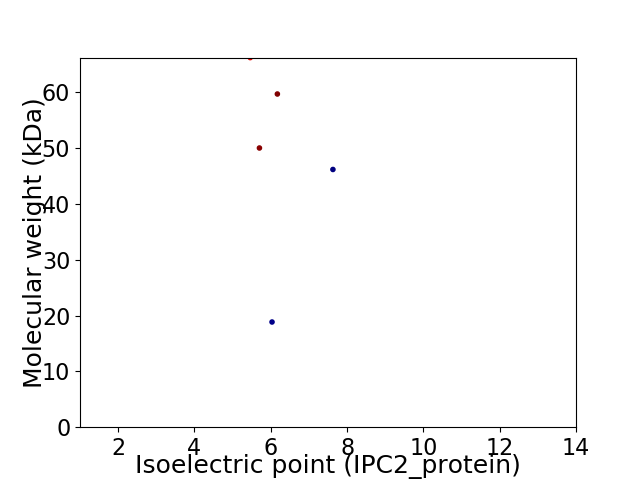
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|B5SPY3|B5SPY3_9PAPI Regulatory protein E2 OS=Sus scrofa papillomavirus 1 OX=446138 GN=E2 PE=3 SV=1
MM1 pKa = 7.71EE2 pKa = 5.68EE3 pKa = 4.35GNSPLEE9 pKa = 4.46GCSGDD14 pKa = 3.72WILVEE19 pKa = 4.39AEE21 pKa = 4.38EE22 pKa = 4.64EE23 pKa = 4.39GGACGGDD30 pKa = 3.44GEE32 pKa = 4.69TDD34 pKa = 3.24SDD36 pKa = 4.34DD37 pKa = 4.64GYY39 pKa = 11.42DD40 pKa = 3.35MVDD43 pKa = 4.99FIDD46 pKa = 4.19DD47 pKa = 3.92TEE49 pKa = 4.56VPQGNSAALYY59 pKa = 9.36HH60 pKa = 6.46AQTLYY65 pKa = 10.01DD66 pKa = 3.72TAVLPQCKK74 pKa = 8.82RR75 pKa = 11.84KK76 pKa = 9.56HH77 pKa = 5.81VNSPKK82 pKa = 10.32VKK84 pKa = 9.39PVCDD88 pKa = 4.3LSPRR92 pKa = 11.84LAAINIGTEE101 pKa = 4.04SQQTAKK107 pKa = 10.35RR108 pKa = 11.84RR109 pKa = 11.84LFVAPPDD116 pKa = 3.59SGYY119 pKa = 11.55GNTLEE124 pKa = 4.18EE125 pKa = 4.97ANTQEE130 pKa = 4.11MGEE133 pKa = 4.35TEE135 pKa = 4.3VPGGNGGEE143 pKa = 4.32GEE145 pKa = 4.34HH146 pKa = 5.97MVAEE150 pKa = 4.36AFGQEE155 pKa = 3.68RR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84AAMLGQFKK166 pKa = 10.41EE167 pKa = 4.0RR168 pKa = 11.84FGISFCDD175 pKa = 3.29LTRR178 pKa = 11.84PFKK181 pKa = 11.09SNKK184 pKa = 5.4TTCGDD189 pKa = 3.35WVVYY193 pKa = 10.05ACGMSDD199 pKa = 3.76ALADD203 pKa = 3.72ATSDD207 pKa = 3.93LLSPHH212 pKa = 7.2CIYY215 pKa = 11.37GNITKK220 pKa = 9.76EE221 pKa = 3.89HH222 pKa = 6.01SALGNSLLMLLRR234 pKa = 11.84FTHH237 pKa = 6.58AKK239 pKa = 10.08NRR241 pKa = 11.84DD242 pKa = 3.61TLTKK246 pKa = 10.73LLGSHH251 pKa = 5.4MTIPAYY257 pKa = 10.67NIVAEE262 pKa = 4.39PPKK265 pKa = 10.18IRR267 pKa = 11.84CPAAAIFWYY276 pKa = 10.17RR277 pKa = 11.84RR278 pKa = 11.84STTQGVTVFGDD289 pKa = 3.27VCEE292 pKa = 4.85WITKK296 pKa = 8.87QVTVGGGEE304 pKa = 4.26CAFEE308 pKa = 4.52LSTMIQWAYY317 pKa = 11.08DD318 pKa = 3.34SDD320 pKa = 4.15LTDD323 pKa = 3.65EE324 pKa = 4.52AQVAYY329 pKa = 9.65EE330 pKa = 4.01YY331 pKa = 11.57AKK333 pKa = 10.98LADD336 pKa = 3.55SDD338 pKa = 4.28RR339 pKa = 11.84NAEE342 pKa = 4.37AFLKK346 pKa = 10.78SNCQAKK352 pKa = 9.52YY353 pKa = 11.04VKK355 pKa = 10.04DD356 pKa = 3.62CCTMVRR362 pKa = 11.84LYY364 pKa = 10.93KK365 pKa = 10.17KK366 pKa = 10.78AEE368 pKa = 3.9MHH370 pKa = 6.13NMTMGQWVRR379 pKa = 11.84KK380 pKa = 8.63QADD383 pKa = 3.81RR384 pKa = 11.84YY385 pKa = 9.6EE386 pKa = 4.86GEE388 pKa = 4.07GNYY391 pKa = 10.03RR392 pKa = 11.84PIVQFLKK399 pKa = 10.28FQGIEE404 pKa = 4.59FIPFMHH410 pKa = 7.07TLAQFLKK417 pKa = 10.65GIPKK421 pKa = 9.96RR422 pKa = 11.84NCLVIHH428 pKa = 6.61GPPNTGKK435 pKa = 10.55SLFTMSLCSFLGGKK449 pKa = 9.53VISFVNSKK457 pKa = 8.57SHH459 pKa = 7.02FWLQPLGEE467 pKa = 4.64CKK469 pKa = 10.17VALLDD474 pKa = 5.14DD475 pKa = 3.97ATRR478 pKa = 11.84PCWDD482 pKa = 3.51YY483 pKa = 11.68FDD485 pKa = 6.11LYY487 pKa = 11.09LRR489 pKa = 11.84NLLDD493 pKa = 4.33GNPVSLDD500 pKa = 3.33AKK502 pKa = 10.13HH503 pKa = 6.26RR504 pKa = 11.84APIQIKK510 pKa = 10.32GPPLLITSNVGISTEE525 pKa = 4.12DD526 pKa = 2.67RR527 pKa = 11.84WKK529 pKa = 10.11YY530 pKa = 8.2LHH532 pKa = 6.71SRR534 pKa = 11.84VRR536 pKa = 11.84CFSFPNTLPLDD547 pKa = 4.05SKK549 pKa = 11.35GDD551 pKa = 3.65PVYY554 pKa = 11.01EE555 pKa = 3.83LTALNWKK562 pKa = 10.19SFFTRR567 pKa = 11.84LWSRR571 pKa = 11.84LDD573 pKa = 3.61LKK575 pKa = 10.85EE576 pKa = 4.3GSHH579 pKa = 6.46GEE581 pKa = 3.68AFTALRR587 pKa = 11.84CTTGPAAGPDD597 pKa = 3.34
MM1 pKa = 7.71EE2 pKa = 5.68EE3 pKa = 4.35GNSPLEE9 pKa = 4.46GCSGDD14 pKa = 3.72WILVEE19 pKa = 4.39AEE21 pKa = 4.38EE22 pKa = 4.64EE23 pKa = 4.39GGACGGDD30 pKa = 3.44GEE32 pKa = 4.69TDD34 pKa = 3.24SDD36 pKa = 4.34DD37 pKa = 4.64GYY39 pKa = 11.42DD40 pKa = 3.35MVDD43 pKa = 4.99FIDD46 pKa = 4.19DD47 pKa = 3.92TEE49 pKa = 4.56VPQGNSAALYY59 pKa = 9.36HH60 pKa = 6.46AQTLYY65 pKa = 10.01DD66 pKa = 3.72TAVLPQCKK74 pKa = 8.82RR75 pKa = 11.84KK76 pKa = 9.56HH77 pKa = 5.81VNSPKK82 pKa = 10.32VKK84 pKa = 9.39PVCDD88 pKa = 4.3LSPRR92 pKa = 11.84LAAINIGTEE101 pKa = 4.04SQQTAKK107 pKa = 10.35RR108 pKa = 11.84RR109 pKa = 11.84LFVAPPDD116 pKa = 3.59SGYY119 pKa = 11.55GNTLEE124 pKa = 4.18EE125 pKa = 4.97ANTQEE130 pKa = 4.11MGEE133 pKa = 4.35TEE135 pKa = 4.3VPGGNGGEE143 pKa = 4.32GEE145 pKa = 4.34HH146 pKa = 5.97MVAEE150 pKa = 4.36AFGQEE155 pKa = 3.68RR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84AAMLGQFKK166 pKa = 10.41EE167 pKa = 4.0RR168 pKa = 11.84FGISFCDD175 pKa = 3.29LTRR178 pKa = 11.84PFKK181 pKa = 11.09SNKK184 pKa = 5.4TTCGDD189 pKa = 3.35WVVYY193 pKa = 10.05ACGMSDD199 pKa = 3.76ALADD203 pKa = 3.72ATSDD207 pKa = 3.93LLSPHH212 pKa = 7.2CIYY215 pKa = 11.37GNITKK220 pKa = 9.76EE221 pKa = 3.89HH222 pKa = 6.01SALGNSLLMLLRR234 pKa = 11.84FTHH237 pKa = 6.58AKK239 pKa = 10.08NRR241 pKa = 11.84DD242 pKa = 3.61TLTKK246 pKa = 10.73LLGSHH251 pKa = 5.4MTIPAYY257 pKa = 10.67NIVAEE262 pKa = 4.39PPKK265 pKa = 10.18IRR267 pKa = 11.84CPAAAIFWYY276 pKa = 10.17RR277 pKa = 11.84RR278 pKa = 11.84STTQGVTVFGDD289 pKa = 3.27VCEE292 pKa = 4.85WITKK296 pKa = 8.87QVTVGGGEE304 pKa = 4.26CAFEE308 pKa = 4.52LSTMIQWAYY317 pKa = 11.08DD318 pKa = 3.34SDD320 pKa = 4.15LTDD323 pKa = 3.65EE324 pKa = 4.52AQVAYY329 pKa = 9.65EE330 pKa = 4.01YY331 pKa = 11.57AKK333 pKa = 10.98LADD336 pKa = 3.55SDD338 pKa = 4.28RR339 pKa = 11.84NAEE342 pKa = 4.37AFLKK346 pKa = 10.78SNCQAKK352 pKa = 9.52YY353 pKa = 11.04VKK355 pKa = 10.04DD356 pKa = 3.62CCTMVRR362 pKa = 11.84LYY364 pKa = 10.93KK365 pKa = 10.17KK366 pKa = 10.78AEE368 pKa = 3.9MHH370 pKa = 6.13NMTMGQWVRR379 pKa = 11.84KK380 pKa = 8.63QADD383 pKa = 3.81RR384 pKa = 11.84YY385 pKa = 9.6EE386 pKa = 4.86GEE388 pKa = 4.07GNYY391 pKa = 10.03RR392 pKa = 11.84PIVQFLKK399 pKa = 10.28FQGIEE404 pKa = 4.59FIPFMHH410 pKa = 7.07TLAQFLKK417 pKa = 10.65GIPKK421 pKa = 9.96RR422 pKa = 11.84NCLVIHH428 pKa = 6.61GPPNTGKK435 pKa = 10.55SLFTMSLCSFLGGKK449 pKa = 9.53VISFVNSKK457 pKa = 8.57SHH459 pKa = 7.02FWLQPLGEE467 pKa = 4.64CKK469 pKa = 10.17VALLDD474 pKa = 5.14DD475 pKa = 3.97ATRR478 pKa = 11.84PCWDD482 pKa = 3.51YY483 pKa = 11.68FDD485 pKa = 6.11LYY487 pKa = 11.09LRR489 pKa = 11.84NLLDD493 pKa = 4.33GNPVSLDD500 pKa = 3.33AKK502 pKa = 10.13HH503 pKa = 6.26RR504 pKa = 11.84APIQIKK510 pKa = 10.32GPPLLITSNVGISTEE525 pKa = 4.12DD526 pKa = 2.67RR527 pKa = 11.84WKK529 pKa = 10.11YY530 pKa = 8.2LHH532 pKa = 6.71SRR534 pKa = 11.84VRR536 pKa = 11.84CFSFPNTLPLDD547 pKa = 4.05SKK549 pKa = 11.35GDD551 pKa = 3.65PVYY554 pKa = 11.01EE555 pKa = 3.83LTALNWKK562 pKa = 10.19SFFTRR567 pKa = 11.84LWSRR571 pKa = 11.84LDD573 pKa = 3.61LKK575 pKa = 10.85EE576 pKa = 4.3GSHH579 pKa = 6.46GEE581 pKa = 3.68AFTALRR587 pKa = 11.84CTTGPAAGPDD597 pKa = 3.34
Molecular weight: 66.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B5SPY4|B5SPY4_9PAPI Minor capsid protein L2 OS=Sus scrofa papillomavirus 1 OX=446138 GN=L2 PE=3 SV=1
MM1 pKa = 7.42EE2 pKa = 5.9RR3 pKa = 11.84LSQRR7 pKa = 11.84LGALQDD13 pKa = 3.55QQLDD17 pKa = 4.36LIEE20 pKa = 5.58KK21 pKa = 9.54DD22 pKa = 3.65SQSIGDD28 pKa = 3.72QLYY31 pKa = 8.17YY32 pKa = 10.19WYY34 pKa = 10.32LQRR37 pKa = 11.84QEE39 pKa = 4.16HH40 pKa = 5.52TLLYY44 pKa = 10.01AARR47 pKa = 11.84KK48 pKa = 9.78KK49 pKa = 10.87GVTSINGQPVPTAQVSHH66 pKa = 6.26SRR68 pKa = 11.84ASQAIEE74 pKa = 3.62MHH76 pKa = 6.79LALQGLGQLYY86 pKa = 10.58AEE88 pKa = 5.17EE89 pKa = 4.77PWTMLQTTWEE99 pKa = 4.59SYY101 pKa = 10.15RR102 pKa = 11.84QTPSGTFKK110 pKa = 10.92KK111 pKa = 10.02GGLSIRR117 pKa = 11.84VQFDD121 pKa = 3.63GEE123 pKa = 4.4KK124 pKa = 10.41DD125 pKa = 3.43NEE127 pKa = 3.98MDD129 pKa = 3.67YY130 pKa = 11.39VLWDD134 pKa = 3.3TVYY137 pKa = 11.64YY138 pKa = 7.09MTGDD142 pKa = 3.53GGWRR146 pKa = 11.84KK147 pKa = 9.96GKK149 pKa = 10.58GGWDD153 pKa = 3.37NTGLYY158 pKa = 10.3YY159 pKa = 10.95LADD162 pKa = 4.05GEE164 pKa = 4.36KK165 pKa = 9.53QHH167 pKa = 6.08YY168 pKa = 10.78VNFKK172 pKa = 9.94TEE174 pKa = 3.74AQKK177 pKa = 10.38FAKK180 pKa = 10.31LGIWKK185 pKa = 8.53VLCTDD190 pKa = 3.34SHH192 pKa = 6.86HH193 pKa = 7.72DD194 pKa = 3.62SDD196 pKa = 5.53LVTSTTTATSTSTQSLSGPCWDD218 pKa = 4.1PHH220 pKa = 6.43PPTTTAEE227 pKa = 4.23STSRR231 pKa = 11.84SPWDD235 pKa = 3.63TPPTEE240 pKa = 3.73TAARR244 pKa = 11.84RR245 pKa = 11.84RR246 pKa = 11.84LATTATPCARR256 pKa = 11.84RR257 pKa = 11.84PQAVPADD264 pKa = 3.99SPSPVPRR271 pKa = 11.84WGPGQGKK278 pKa = 8.99HH279 pKa = 6.27PSTPDD284 pKa = 2.86ATAKK288 pKa = 8.37PTAAPAATNTTTYY301 pKa = 7.03TTTAGRR307 pKa = 11.84GSIGPQCLCDD317 pKa = 4.18CDD319 pKa = 5.63PNTTWGSEE327 pKa = 4.11SQAGVPLTAPLVVLKK342 pKa = 10.8GDD344 pKa = 3.92PNQLKK349 pKa = 10.27CYY351 pKa = 10.16RR352 pKa = 11.84YY353 pKa = 9.01RR354 pKa = 11.84LKK356 pKa = 10.54KK357 pKa = 10.19GRR359 pKa = 11.84RR360 pKa = 11.84KK361 pKa = 9.93SYY363 pKa = 9.75RR364 pKa = 11.84HH365 pKa = 6.83CSTTWQWVGCDD376 pKa = 3.28GVSRR380 pKa = 11.84TGKK383 pKa = 9.83HH384 pKa = 5.55LLCVSFDD391 pKa = 3.55NSAQRR396 pKa = 11.84EE397 pKa = 4.36EE398 pKa = 4.36FLLTAQLPQGVTATRR413 pKa = 11.84ADD415 pKa = 3.78FPLL418 pKa = 5.26
MM1 pKa = 7.42EE2 pKa = 5.9RR3 pKa = 11.84LSQRR7 pKa = 11.84LGALQDD13 pKa = 3.55QQLDD17 pKa = 4.36LIEE20 pKa = 5.58KK21 pKa = 9.54DD22 pKa = 3.65SQSIGDD28 pKa = 3.72QLYY31 pKa = 8.17YY32 pKa = 10.19WYY34 pKa = 10.32LQRR37 pKa = 11.84QEE39 pKa = 4.16HH40 pKa = 5.52TLLYY44 pKa = 10.01AARR47 pKa = 11.84KK48 pKa = 9.78KK49 pKa = 10.87GVTSINGQPVPTAQVSHH66 pKa = 6.26SRR68 pKa = 11.84ASQAIEE74 pKa = 3.62MHH76 pKa = 6.79LALQGLGQLYY86 pKa = 10.58AEE88 pKa = 5.17EE89 pKa = 4.77PWTMLQTTWEE99 pKa = 4.59SYY101 pKa = 10.15RR102 pKa = 11.84QTPSGTFKK110 pKa = 10.92KK111 pKa = 10.02GGLSIRR117 pKa = 11.84VQFDD121 pKa = 3.63GEE123 pKa = 4.4KK124 pKa = 10.41DD125 pKa = 3.43NEE127 pKa = 3.98MDD129 pKa = 3.67YY130 pKa = 11.39VLWDD134 pKa = 3.3TVYY137 pKa = 11.64YY138 pKa = 7.09MTGDD142 pKa = 3.53GGWRR146 pKa = 11.84KK147 pKa = 9.96GKK149 pKa = 10.58GGWDD153 pKa = 3.37NTGLYY158 pKa = 10.3YY159 pKa = 10.95LADD162 pKa = 4.05GEE164 pKa = 4.36KK165 pKa = 9.53QHH167 pKa = 6.08YY168 pKa = 10.78VNFKK172 pKa = 9.94TEE174 pKa = 3.74AQKK177 pKa = 10.38FAKK180 pKa = 10.31LGIWKK185 pKa = 8.53VLCTDD190 pKa = 3.34SHH192 pKa = 6.86HH193 pKa = 7.72DD194 pKa = 3.62SDD196 pKa = 5.53LVTSTTTATSTSTQSLSGPCWDD218 pKa = 4.1PHH220 pKa = 6.43PPTTTAEE227 pKa = 4.23STSRR231 pKa = 11.84SPWDD235 pKa = 3.63TPPTEE240 pKa = 3.73TAARR244 pKa = 11.84RR245 pKa = 11.84RR246 pKa = 11.84LATTATPCARR256 pKa = 11.84RR257 pKa = 11.84PQAVPADD264 pKa = 3.99SPSPVPRR271 pKa = 11.84WGPGQGKK278 pKa = 8.99HH279 pKa = 6.27PSTPDD284 pKa = 2.86ATAKK288 pKa = 8.37PTAAPAATNTTTYY301 pKa = 7.03TTTAGRR307 pKa = 11.84GSIGPQCLCDD317 pKa = 4.18CDD319 pKa = 5.63PNTTWGSEE327 pKa = 4.11SQAGVPLTAPLVVLKK342 pKa = 10.8GDD344 pKa = 3.92PNQLKK349 pKa = 10.27CYY351 pKa = 10.16RR352 pKa = 11.84YY353 pKa = 9.01RR354 pKa = 11.84LKK356 pKa = 10.54KK357 pKa = 10.19GRR359 pKa = 11.84RR360 pKa = 11.84KK361 pKa = 9.93SYY363 pKa = 9.75RR364 pKa = 11.84HH365 pKa = 6.83CSTTWQWVGCDD376 pKa = 3.28GVSRR380 pKa = 11.84TGKK383 pKa = 9.83HH384 pKa = 5.55LLCVSFDD391 pKa = 3.55NSAQRR396 pKa = 11.84EE397 pKa = 4.36EE398 pKa = 4.36FLLTAQLPQGVTATRR413 pKa = 11.84ADD415 pKa = 3.78FPLL418 pKa = 5.26
Molecular weight: 46.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2185 |
165 |
597 |
437.0 |
48.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.499 ± 0.661 | 2.38 ± 0.603 |
6.224 ± 0.166 | 4.805 ± 0.519 |
4.027 ± 0.567 | 8.65 ± 1.077 |
2.288 ± 0.057 | 3.432 ± 0.528 |
4.119 ± 0.753 | 8.741 ± 0.417 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.739 ± 0.29 | 3.341 ± 0.731 |
7.506 ± 1.15 | 4.165 ± 0.732 |
6.178 ± 0.595 | 7.048 ± 0.391 |
8.146 ± 0.88 | 5.995 ± 0.713 |
1.831 ± 0.413 | 2.883 ± 0.41 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
