
Synechococcus phage S-WAM2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Cymopoleiavirus; Synechococcus virus SWAM2
Average proteome isoelectric point is 5.74
Get precalculated fractions of proteins
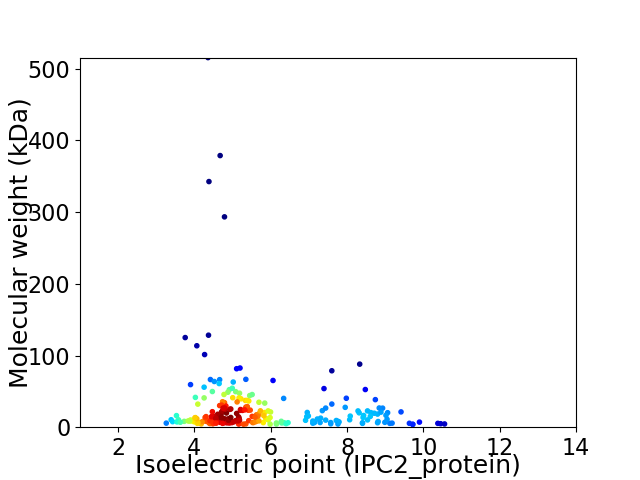
Virtual 2D-PAGE plot for 229 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D8KSY2|A0A1D8KSY2_9CAUD Uncharacterized protein OS=Synechococcus phage S-WAM2 OX=1815522 GN=P29B0810_095 PE=4 SV=1
MM1 pKa = 7.39GFYY4 pKa = 10.6RR5 pKa = 11.84KK6 pKa = 7.73TLGVLSAASTDD17 pKa = 3.28KK18 pKa = 11.24DD19 pKa = 3.44IVDD22 pKa = 4.7FALTGTVFFTDD33 pKa = 3.14VNLRR37 pKa = 11.84KK38 pKa = 10.18YY39 pKa = 10.57FIADD43 pKa = 3.57DD44 pKa = 3.99NVVISSPGSNYY55 pKa = 10.37SVGDD59 pKa = 3.84YY60 pKa = 11.15VSVPLLGGTGSGAVADD76 pKa = 3.95IGVNDD81 pKa = 4.03WVGSITNAGSGYY93 pKa = 9.02TEE95 pKa = 3.89GTYY98 pKa = 10.28TSLYY102 pKa = 9.13PYY104 pKa = 9.91VISGAGNGDD113 pKa = 3.82ALVAISVNEE122 pKa = 3.88NGNISSFTVEE132 pKa = 3.95QYY134 pKa = 9.51GTGYY138 pKa = 7.89TQGTVLGLYY147 pKa = 10.05GATTSATTLSTVDD160 pKa = 3.97FNVTVSDD167 pKa = 3.64TSSILVNSAVTGTGIAEE184 pKa = 4.35GTTVASVLDD193 pKa = 3.92GTTVEE198 pKa = 4.35LSAYY202 pKa = 9.53PEE204 pKa = 3.85IDD206 pKa = 3.33GASTLTFTPSYY217 pKa = 10.81GGGNGFQFTISAINAVGTVTITDD240 pKa = 3.83GGNGYY245 pKa = 10.12AAGDD249 pKa = 3.94TLTVADD255 pKa = 4.94ASLSVPIEE263 pKa = 3.99YY264 pKa = 10.2VVSDD268 pKa = 4.15LSVQRR273 pKa = 11.84LTFVEE278 pKa = 4.92TIPVGTFSIGGTLEE292 pKa = 3.63ISTLDD297 pKa = 3.21GTQYY301 pKa = 11.14NIVEE305 pKa = 4.37IEE307 pKa = 4.12TSGGNITSVVLRR319 pKa = 11.84DD320 pKa = 3.59SFFGATEE327 pKa = 3.75TLAYY331 pKa = 9.96NGSATPIYY339 pKa = 9.16TVDD342 pKa = 3.14VATSEE347 pKa = 3.99FRR349 pKa = 11.84AGIDD353 pKa = 2.98IGDD356 pKa = 3.89GNGVQFTPNLTLYY369 pKa = 10.72EE370 pKa = 4.32DD371 pKa = 3.27ATYY374 pKa = 10.89EE375 pKa = 4.04FTYY378 pKa = 10.65SGSNTFALSQTQDD391 pKa = 3.06GPGEE395 pKa = 4.15YY396 pKa = 10.37APSTVVRR403 pKa = 11.84DD404 pKa = 4.01TEE406 pKa = 4.45NNTLTVSVTDD416 pKa = 4.31SYY418 pKa = 11.72PSTLYY423 pKa = 10.88YY424 pKa = 10.73FSTDD428 pKa = 2.85NPNYY432 pKa = 9.92GGSSSITIDD441 pKa = 3.72PNNPSPPGTGFQLLVNTVTILDD463 pKa = 4.54SIALDD468 pKa = 4.09IIDD471 pKa = 4.61GSVTALDD478 pKa = 4.21VITTNLTATTGTVSTLTSTSGDD500 pKa = 2.86ITTLKK505 pKa = 9.03ITSLSDD511 pKa = 3.0KK512 pKa = 10.73GSGIAVTTGGSTNITLTPGNSLNVGGALSIEE543 pKa = 4.47SATGNLTTSGVLKK556 pKa = 10.05STGSFNSNDD565 pKa = 3.17KK566 pKa = 11.11LLISEE571 pKa = 4.62NSISSLGTSDD581 pKa = 4.8VVLAPIANTNAKK593 pKa = 10.09VSGTMSLIIPSGDD606 pKa = 2.61ISQRR610 pKa = 11.84PQGAKK615 pKa = 10.22AEE617 pKa = 4.3SGSIRR622 pKa = 11.84FNTEE626 pKa = 3.29TQQYY630 pKa = 9.2EE631 pKa = 5.01GYY633 pKa = 10.54NGIAAAWSSLGGVRR647 pKa = 11.84DD648 pKa = 3.63VDD650 pKa = 3.62GNTYY654 pKa = 10.3ILAEE658 pKa = 4.45EE659 pKa = 4.09FTGANDD665 pKa = 3.31NTFWFYY671 pKa = 11.73NGGTNSLTISQSEE684 pKa = 4.32INLKK688 pKa = 10.03SANKK692 pKa = 9.66FKK694 pKa = 10.56STDD697 pKa = 3.39VTSYY701 pKa = 10.51TPWAGNTYY709 pKa = 10.21YY710 pKa = 11.38GLGEE714 pKa = 5.27LLYY717 pKa = 10.55QDD719 pKa = 4.16LNVYY723 pKa = 9.12EE724 pKa = 4.25VTVAGLSDD732 pKa = 4.44TIAPTHH738 pKa = 5.46EE739 pKa = 4.45TGAVTANGGTPTIAGAIAAFTVSTPSGNNAVGTYY773 pKa = 10.03SYY775 pKa = 11.58DD776 pKa = 3.42DD777 pKa = 3.95TYY779 pKa = 11.9GSAQFEE785 pKa = 4.99IIFQLGTVEE794 pKa = 5.16VNLLSQGSGGYY805 pKa = 7.8STSGGGVDD813 pKa = 4.1NVITILGSLIGGTDD827 pKa = 3.49GVDD830 pKa = 3.47DD831 pKa = 4.03LTITITEE838 pKa = 3.89IAVALEE844 pKa = 3.99LTWYY848 pKa = 7.53GTTAGNIEE856 pKa = 4.2FEE858 pKa = 4.72DD859 pKa = 3.68VNEE862 pKa = 4.46VIFTDD867 pKa = 3.99STIHH871 pKa = 6.75ASNTISGRR879 pKa = 11.84NLLITGGEE887 pKa = 4.0IEE889 pKa = 4.45GDD891 pKa = 3.24GDD893 pKa = 4.3LSIKK897 pKa = 10.47IPDD900 pKa = 3.75GSNLVVTATGSLAVPVGDD918 pKa = 3.29NLQRR922 pKa = 11.84GTPVAGSIRR931 pKa = 11.84YY932 pKa = 9.14NNEE935 pKa = 2.92INQYY939 pKa = 9.94EE940 pKa = 5.04GYY942 pKa = 10.93NSAASNWSSLGGVRR956 pKa = 11.84DD957 pKa = 3.63VDD959 pKa = 3.61GNTYY963 pKa = 10.37IIPEE967 pKa = 4.15SSAGANEE974 pKa = 4.39NILYY978 pKa = 9.77FYY980 pKa = 11.37NNDD983 pKa = 3.69DD984 pKa = 3.37NTLQVTQSQVLFGTIDD1000 pKa = 4.01SISSTSNALNINVEE1014 pKa = 4.05NVTFNNLHH1022 pKa = 6.2SGIDD1026 pKa = 3.59VSDD1029 pKa = 3.44ATTTLVYY1036 pKa = 10.83SSVNNLDD1043 pKa = 3.91FGLSTGLTIDD1053 pKa = 3.36TVLRR1057 pKa = 11.84LTDD1060 pKa = 3.51AGEE1063 pKa = 3.83IFLNKK1068 pKa = 10.33GYY1070 pKa = 8.6GTGVFEE1076 pKa = 4.23GVKK1079 pKa = 10.6FIDD1082 pKa = 3.71AFLKK1086 pKa = 10.09TFEE1089 pKa = 5.14LDD1091 pKa = 3.34DD1092 pKa = 3.98VQIKK1096 pKa = 9.58TDD1098 pKa = 3.94DD1099 pKa = 3.59VVLTKK1104 pKa = 10.16GTTDD1108 pKa = 2.78TGTVVLYY1115 pKa = 10.76DD1116 pKa = 3.55PVEE1119 pKa = 4.28AKK1121 pKa = 9.9SSKK1124 pKa = 10.83VIVTAYY1130 pKa = 7.97NTTTEE1135 pKa = 4.16DD1136 pKa = 3.31VHH1138 pKa = 7.28TEE1140 pKa = 4.13EE1141 pKa = 4.95ISVIVKK1147 pKa = 10.58GSDD1150 pKa = 3.11LYY1152 pKa = 10.08TVEE1155 pKa = 4.16YY1156 pKa = 8.53GTNKK1160 pKa = 6.72TTNLFAAVVDD1170 pKa = 4.92LNATGKK1176 pKa = 9.85VRR1178 pKa = 11.84LSLALDD1184 pKa = 3.33SGIATGEE1191 pKa = 3.96IVNITVVRR1199 pKa = 11.84TNVKK1203 pKa = 10.03KK1204 pKa = 10.88
MM1 pKa = 7.39GFYY4 pKa = 10.6RR5 pKa = 11.84KK6 pKa = 7.73TLGVLSAASTDD17 pKa = 3.28KK18 pKa = 11.24DD19 pKa = 3.44IVDD22 pKa = 4.7FALTGTVFFTDD33 pKa = 3.14VNLRR37 pKa = 11.84KK38 pKa = 10.18YY39 pKa = 10.57FIADD43 pKa = 3.57DD44 pKa = 3.99NVVISSPGSNYY55 pKa = 10.37SVGDD59 pKa = 3.84YY60 pKa = 11.15VSVPLLGGTGSGAVADD76 pKa = 3.95IGVNDD81 pKa = 4.03WVGSITNAGSGYY93 pKa = 9.02TEE95 pKa = 3.89GTYY98 pKa = 10.28TSLYY102 pKa = 9.13PYY104 pKa = 9.91VISGAGNGDD113 pKa = 3.82ALVAISVNEE122 pKa = 3.88NGNISSFTVEE132 pKa = 3.95QYY134 pKa = 9.51GTGYY138 pKa = 7.89TQGTVLGLYY147 pKa = 10.05GATTSATTLSTVDD160 pKa = 3.97FNVTVSDD167 pKa = 3.64TSSILVNSAVTGTGIAEE184 pKa = 4.35GTTVASVLDD193 pKa = 3.92GTTVEE198 pKa = 4.35LSAYY202 pKa = 9.53PEE204 pKa = 3.85IDD206 pKa = 3.33GASTLTFTPSYY217 pKa = 10.81GGGNGFQFTISAINAVGTVTITDD240 pKa = 3.83GGNGYY245 pKa = 10.12AAGDD249 pKa = 3.94TLTVADD255 pKa = 4.94ASLSVPIEE263 pKa = 3.99YY264 pKa = 10.2VVSDD268 pKa = 4.15LSVQRR273 pKa = 11.84LTFVEE278 pKa = 4.92TIPVGTFSIGGTLEE292 pKa = 3.63ISTLDD297 pKa = 3.21GTQYY301 pKa = 11.14NIVEE305 pKa = 4.37IEE307 pKa = 4.12TSGGNITSVVLRR319 pKa = 11.84DD320 pKa = 3.59SFFGATEE327 pKa = 3.75TLAYY331 pKa = 9.96NGSATPIYY339 pKa = 9.16TVDD342 pKa = 3.14VATSEE347 pKa = 3.99FRR349 pKa = 11.84AGIDD353 pKa = 2.98IGDD356 pKa = 3.89GNGVQFTPNLTLYY369 pKa = 10.72EE370 pKa = 4.32DD371 pKa = 3.27ATYY374 pKa = 10.89EE375 pKa = 4.04FTYY378 pKa = 10.65SGSNTFALSQTQDD391 pKa = 3.06GPGEE395 pKa = 4.15YY396 pKa = 10.37APSTVVRR403 pKa = 11.84DD404 pKa = 4.01TEE406 pKa = 4.45NNTLTVSVTDD416 pKa = 4.31SYY418 pKa = 11.72PSTLYY423 pKa = 10.88YY424 pKa = 10.73FSTDD428 pKa = 2.85NPNYY432 pKa = 9.92GGSSSITIDD441 pKa = 3.72PNNPSPPGTGFQLLVNTVTILDD463 pKa = 4.54SIALDD468 pKa = 4.09IIDD471 pKa = 4.61GSVTALDD478 pKa = 4.21VITTNLTATTGTVSTLTSTSGDD500 pKa = 2.86ITTLKK505 pKa = 9.03ITSLSDD511 pKa = 3.0KK512 pKa = 10.73GSGIAVTTGGSTNITLTPGNSLNVGGALSIEE543 pKa = 4.47SATGNLTTSGVLKK556 pKa = 10.05STGSFNSNDD565 pKa = 3.17KK566 pKa = 11.11LLISEE571 pKa = 4.62NSISSLGTSDD581 pKa = 4.8VVLAPIANTNAKK593 pKa = 10.09VSGTMSLIIPSGDD606 pKa = 2.61ISQRR610 pKa = 11.84PQGAKK615 pKa = 10.22AEE617 pKa = 4.3SGSIRR622 pKa = 11.84FNTEE626 pKa = 3.29TQQYY630 pKa = 9.2EE631 pKa = 5.01GYY633 pKa = 10.54NGIAAAWSSLGGVRR647 pKa = 11.84DD648 pKa = 3.63VDD650 pKa = 3.62GNTYY654 pKa = 10.3ILAEE658 pKa = 4.45EE659 pKa = 4.09FTGANDD665 pKa = 3.31NTFWFYY671 pKa = 11.73NGGTNSLTISQSEE684 pKa = 4.32INLKK688 pKa = 10.03SANKK692 pKa = 9.66FKK694 pKa = 10.56STDD697 pKa = 3.39VTSYY701 pKa = 10.51TPWAGNTYY709 pKa = 10.21YY710 pKa = 11.38GLGEE714 pKa = 5.27LLYY717 pKa = 10.55QDD719 pKa = 4.16LNVYY723 pKa = 9.12EE724 pKa = 4.25VTVAGLSDD732 pKa = 4.44TIAPTHH738 pKa = 5.46EE739 pKa = 4.45TGAVTANGGTPTIAGAIAAFTVSTPSGNNAVGTYY773 pKa = 10.03SYY775 pKa = 11.58DD776 pKa = 3.42DD777 pKa = 3.95TYY779 pKa = 11.9GSAQFEE785 pKa = 4.99IIFQLGTVEE794 pKa = 5.16VNLLSQGSGGYY805 pKa = 7.8STSGGGVDD813 pKa = 4.1NVITILGSLIGGTDD827 pKa = 3.49GVDD830 pKa = 3.47DD831 pKa = 4.03LTITITEE838 pKa = 3.89IAVALEE844 pKa = 3.99LTWYY848 pKa = 7.53GTTAGNIEE856 pKa = 4.2FEE858 pKa = 4.72DD859 pKa = 3.68VNEE862 pKa = 4.46VIFTDD867 pKa = 3.99STIHH871 pKa = 6.75ASNTISGRR879 pKa = 11.84NLLITGGEE887 pKa = 4.0IEE889 pKa = 4.45GDD891 pKa = 3.24GDD893 pKa = 4.3LSIKK897 pKa = 10.47IPDD900 pKa = 3.75GSNLVVTATGSLAVPVGDD918 pKa = 3.29NLQRR922 pKa = 11.84GTPVAGSIRR931 pKa = 11.84YY932 pKa = 9.14NNEE935 pKa = 2.92INQYY939 pKa = 9.94EE940 pKa = 5.04GYY942 pKa = 10.93NSAASNWSSLGGVRR956 pKa = 11.84DD957 pKa = 3.63VDD959 pKa = 3.61GNTYY963 pKa = 10.37IIPEE967 pKa = 4.15SSAGANEE974 pKa = 4.39NILYY978 pKa = 9.77FYY980 pKa = 11.37NNDD983 pKa = 3.69DD984 pKa = 3.37NTLQVTQSQVLFGTIDD1000 pKa = 4.01SISSTSNALNINVEE1014 pKa = 4.05NVTFNNLHH1022 pKa = 6.2SGIDD1026 pKa = 3.59VSDD1029 pKa = 3.44ATTTLVYY1036 pKa = 10.83SSVNNLDD1043 pKa = 3.91FGLSTGLTIDD1053 pKa = 3.36TVLRR1057 pKa = 11.84LTDD1060 pKa = 3.51AGEE1063 pKa = 3.83IFLNKK1068 pKa = 10.33GYY1070 pKa = 8.6GTGVFEE1076 pKa = 4.23GVKK1079 pKa = 10.6FIDD1082 pKa = 3.71AFLKK1086 pKa = 10.09TFEE1089 pKa = 5.14LDD1091 pKa = 3.34DD1092 pKa = 3.98VQIKK1096 pKa = 9.58TDD1098 pKa = 3.94DD1099 pKa = 3.59VVLTKK1104 pKa = 10.16GTTDD1108 pKa = 2.78TGTVVLYY1115 pKa = 10.76DD1116 pKa = 3.55PVEE1119 pKa = 4.28AKK1121 pKa = 9.9SSKK1124 pKa = 10.83VIVTAYY1130 pKa = 7.97NTTTEE1135 pKa = 4.16DD1136 pKa = 3.31VHH1138 pKa = 7.28TEE1140 pKa = 4.13EE1141 pKa = 4.95ISVIVKK1147 pKa = 10.58GSDD1150 pKa = 3.11LYY1152 pKa = 10.08TVEE1155 pKa = 4.16YY1156 pKa = 8.53GTNKK1160 pKa = 6.72TTNLFAAVVDD1170 pKa = 4.92LNATGKK1176 pKa = 9.85VRR1178 pKa = 11.84LSLALDD1184 pKa = 3.33SGIATGEE1191 pKa = 3.96IVNITVVRR1199 pKa = 11.84TNVKK1203 pKa = 10.03KK1204 pKa = 10.88
Molecular weight: 125.02 kDa
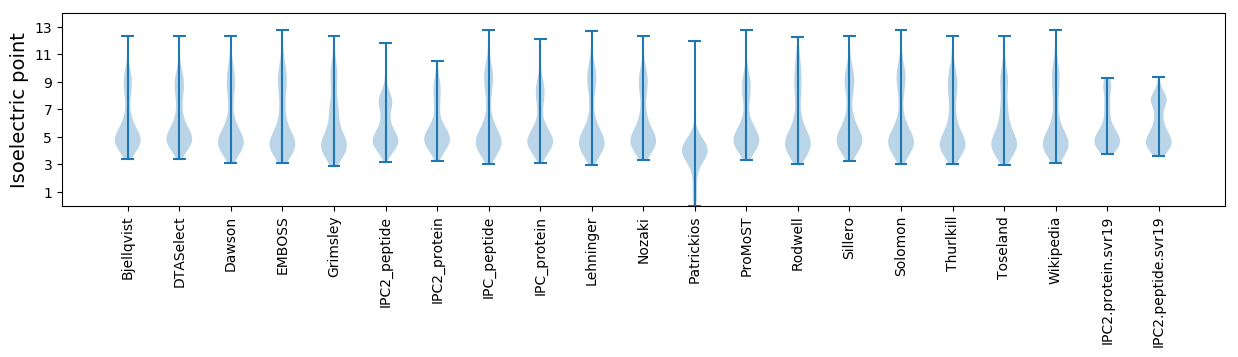
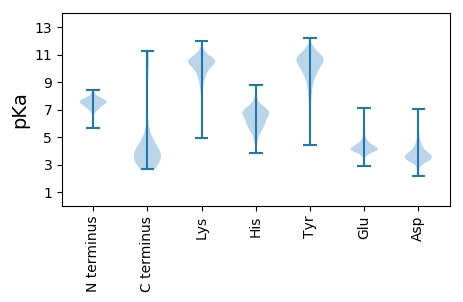
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D8KSU6|A0A1D8KSU6_9CAUD Carbon metabolic regulator OS=Synechococcus phage S-WAM2 OX=1815522 GN=P29B0810_011 PE=4 SV=1
MM1 pKa = 8.12RR2 pKa = 11.84SGQPDD7 pKa = 3.65PSSAGNHH14 pKa = 5.48SASKK18 pKa = 10.47FSRR21 pKa = 11.84KK22 pKa = 7.49QLCSNLFSQLPLLHH36 pKa = 6.47PLWRR40 pKa = 11.84PLLWQVPMM48 pKa = 4.91
MM1 pKa = 8.12RR2 pKa = 11.84SGQPDD7 pKa = 3.65PSSAGNHH14 pKa = 5.48SASKK18 pKa = 10.47FSRR21 pKa = 11.84KK22 pKa = 7.49QLCSNLFSQLPLLHH36 pKa = 6.47PLWRR40 pKa = 11.84PLLWQVPMM48 pKa = 4.91
Molecular weight: 5.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
57339 |
37 |
4759 |
250.4 |
27.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.321 ± 0.269 | 0.869 ± 0.079 |
6.639 ± 0.167 | 6.17 ± 0.285 |
4.301 ± 0.113 | 7.862 ± 0.369 |
1.428 ± 0.111 | 6.219 ± 0.193 |
5.467 ± 0.333 | 7.135 ± 0.157 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.913 ± 0.198 | 5.972 ± 0.178 |
3.938 ± 0.111 | 3.697 ± 0.105 |
3.929 ± 0.111 | 7.021 ± 0.274 |
7.728 ± 0.333 | 6.807 ± 0.177 |
1.151 ± 0.086 | 4.432 ± 0.111 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
