
Faeces associated gemycircularvirus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus geras2; Gerygone associated gemycircularvirus 2
Average proteome isoelectric point is 7.49
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|T1YRW5|T1YRW5_9VIRU Capsid protein OS=Faeces associated gemycircularvirus 5 OX=1391034 PE=4 SV=1
MM1 pKa = 7.34TFILNARR8 pKa = 11.84YY9 pKa = 8.98FLVTYY14 pKa = 7.78PQSDD18 pKa = 4.04GLDD21 pKa = 2.95EE22 pKa = 4.22WAVNDD27 pKa = 3.93HH28 pKa = 6.47FGSLGAEE35 pKa = 3.91CIVARR40 pKa = 11.84EE41 pKa = 3.93NHH43 pKa = 5.82AVRR46 pKa = 11.84GTHH49 pKa = 6.25LHH51 pKa = 5.32VFCDD55 pKa = 4.88FGRR58 pKa = 11.84KK59 pKa = 8.2FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84ADD66 pKa = 3.04IFDD69 pKa = 3.3VGGFHH74 pKa = 7.62PNIEE78 pKa = 3.93RR79 pKa = 11.84SKK81 pKa = 9.8RR82 pKa = 11.84NPRR85 pKa = 11.84KK86 pKa = 9.72GALYY90 pKa = 10.12ACKK93 pKa = 10.51DD94 pKa = 3.31GDD96 pKa = 3.9IVAGGLDD103 pKa = 3.83VPGLATSIVSAAQDD117 pKa = 3.39PGATLVCAEE126 pKa = 4.19SQRR129 pKa = 11.84EE130 pKa = 4.04FFEE133 pKa = 4.36LAEE136 pKa = 5.66DD137 pKa = 3.75ICPWDD142 pKa = 5.07LITKK146 pKa = 8.77FGSMHH151 pKa = 7.37AYY153 pKa = 10.05AKK155 pKa = 9.95WKK157 pKa = 10.06FPEE160 pKa = 4.06LAEE163 pKa = 4.64PYY165 pKa = 9.08EE166 pKa = 4.38NPAGFTLADD175 pKa = 3.64GAFPDD180 pKa = 3.96LVSWRR185 pKa = 11.84TGALEE190 pKa = 3.52HH191 pKa = 6.61SGRR194 pKa = 11.84IKK196 pKa = 10.88SLVLIGDD203 pKa = 4.37ALTGKK208 pKa = 6.14TTWARR213 pKa = 11.84MLGNHH218 pKa = 7.21LYY220 pKa = 9.94MKK222 pKa = 10.07EE223 pKa = 3.82RR224 pKa = 11.84YY225 pKa = 8.1NAKK228 pKa = 9.6QASLADD234 pKa = 3.69GVDD237 pKa = 3.29YY238 pKa = 11.28GVIDD242 pKa = 5.66DD243 pKa = 3.81ISGGIKK249 pKa = 10.11YY250 pKa = 9.94FPHH253 pKa = 5.57WKK255 pKa = 9.37SWFGGQPHH263 pKa = 6.38IQVKK267 pKa = 9.99ILYY270 pKa = 9.57KK271 pKa = 10.14DD272 pKa = 3.3EE273 pKa = 5.6RR274 pKa = 11.84LVKK277 pKa = 9.28WGKK280 pKa = 8.58PLIWIANRR288 pKa = 11.84DD289 pKa = 3.54PRR291 pKa = 11.84DD292 pKa = 3.29QLRR295 pKa = 11.84DD296 pKa = 3.62MVSRR300 pKa = 11.84DD301 pKa = 3.53YY302 pKa = 11.67SEE304 pKa = 4.38EE305 pKa = 3.73QCNNDD310 pKa = 3.83VYY312 pKa = 10.91WMEE315 pKa = 4.11GNAIFVDD322 pKa = 3.97IRR324 pKa = 11.84QSSIISHH331 pKa = 6.89ANTEE335 pKa = 4.12
MM1 pKa = 7.34TFILNARR8 pKa = 11.84YY9 pKa = 8.98FLVTYY14 pKa = 7.78PQSDD18 pKa = 4.04GLDD21 pKa = 2.95EE22 pKa = 4.22WAVNDD27 pKa = 3.93HH28 pKa = 6.47FGSLGAEE35 pKa = 3.91CIVARR40 pKa = 11.84EE41 pKa = 3.93NHH43 pKa = 5.82AVRR46 pKa = 11.84GTHH49 pKa = 6.25LHH51 pKa = 5.32VFCDD55 pKa = 4.88FGRR58 pKa = 11.84KK59 pKa = 8.2FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84ADD66 pKa = 3.04IFDD69 pKa = 3.3VGGFHH74 pKa = 7.62PNIEE78 pKa = 3.93RR79 pKa = 11.84SKK81 pKa = 9.8RR82 pKa = 11.84NPRR85 pKa = 11.84KK86 pKa = 9.72GALYY90 pKa = 10.12ACKK93 pKa = 10.51DD94 pKa = 3.31GDD96 pKa = 3.9IVAGGLDD103 pKa = 3.83VPGLATSIVSAAQDD117 pKa = 3.39PGATLVCAEE126 pKa = 4.19SQRR129 pKa = 11.84EE130 pKa = 4.04FFEE133 pKa = 4.36LAEE136 pKa = 5.66DD137 pKa = 3.75ICPWDD142 pKa = 5.07LITKK146 pKa = 8.77FGSMHH151 pKa = 7.37AYY153 pKa = 10.05AKK155 pKa = 9.95WKK157 pKa = 10.06FPEE160 pKa = 4.06LAEE163 pKa = 4.64PYY165 pKa = 9.08EE166 pKa = 4.38NPAGFTLADD175 pKa = 3.64GAFPDD180 pKa = 3.96LVSWRR185 pKa = 11.84TGALEE190 pKa = 3.52HH191 pKa = 6.61SGRR194 pKa = 11.84IKK196 pKa = 10.88SLVLIGDD203 pKa = 4.37ALTGKK208 pKa = 6.14TTWARR213 pKa = 11.84MLGNHH218 pKa = 7.21LYY220 pKa = 9.94MKK222 pKa = 10.07EE223 pKa = 3.82RR224 pKa = 11.84YY225 pKa = 8.1NAKK228 pKa = 9.6QASLADD234 pKa = 3.69GVDD237 pKa = 3.29YY238 pKa = 11.28GVIDD242 pKa = 5.66DD243 pKa = 3.81ISGGIKK249 pKa = 10.11YY250 pKa = 9.94FPHH253 pKa = 5.57WKK255 pKa = 9.37SWFGGQPHH263 pKa = 6.38IQVKK267 pKa = 9.99ILYY270 pKa = 9.57KK271 pKa = 10.14DD272 pKa = 3.3EE273 pKa = 5.6RR274 pKa = 11.84LVKK277 pKa = 9.28WGKK280 pKa = 8.58PLIWIANRR288 pKa = 11.84DD289 pKa = 3.54PRR291 pKa = 11.84DD292 pKa = 3.29QLRR295 pKa = 11.84DD296 pKa = 3.62MVSRR300 pKa = 11.84DD301 pKa = 3.53YY302 pKa = 11.67SEE304 pKa = 4.38EE305 pKa = 3.73QCNNDD310 pKa = 3.83VYY312 pKa = 10.91WMEE315 pKa = 4.11GNAIFVDD322 pKa = 3.97IRR324 pKa = 11.84QSSIISHH331 pKa = 6.89ANTEE335 pKa = 4.12
Molecular weight: 37.77 kDa
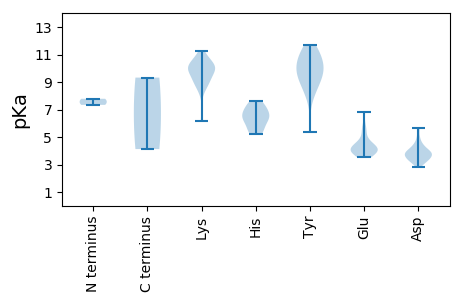
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T1YRW5|T1YRW5_9VIRU Capsid protein OS=Faeces associated gemycircularvirus 5 OX=1391034 PE=4 SV=1
MM1 pKa = 7.78AYY3 pKa = 9.79RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 10.07SYY8 pKa = 10.46SSRR11 pKa = 11.84ARR13 pKa = 11.84RR14 pKa = 11.84PIGRR18 pKa = 11.84SAKK21 pKa = 10.01RR22 pKa = 11.84GGARR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 5.34PTKK32 pKa = 10.21KK33 pKa = 8.56RR34 pKa = 11.84TYY36 pKa = 8.62RR37 pKa = 11.84KK38 pKa = 9.1NRR40 pKa = 11.84AMSTRR45 pKa = 11.84TILNKK50 pKa = 9.28TSRR53 pKa = 11.84KK54 pKa = 8.91KK55 pKa = 10.61RR56 pKa = 11.84NGMLTWANTSTSGTSIPNVATNYY79 pKa = 10.18FLTGNLGGRR88 pKa = 11.84SIWCATAQDD97 pKa = 4.27LTVQDD102 pKa = 4.27GSFGTVALAAARR114 pKa = 11.84TSTSCYY120 pKa = 8.69MKK122 pKa = 10.59GLSEE126 pKa = 5.06HH127 pKa = 6.62IRR129 pKa = 11.84IQTSTGLPWFWRR141 pKa = 11.84RR142 pKa = 11.84ICFTLKK148 pKa = 10.67GDD150 pKa = 3.82DD151 pKa = 3.83LFRR154 pKa = 11.84FSTSDD159 pKa = 3.31TPTSTSNRR167 pKa = 11.84YY168 pKa = 9.6LDD170 pKa = 3.69TTGGMQRR177 pKa = 11.84LFFNEE182 pKa = 3.79LNNAQSNTINNHH194 pKa = 5.19DD195 pKa = 4.5AIIFKK200 pKa = 9.9GAKK203 pKa = 9.23GIDD206 pKa = 3.34WIDD209 pKa = 4.2YY210 pKa = 9.29IVAPIDD216 pKa = 3.37TSRR219 pKa = 11.84INLKK223 pKa = 9.81YY224 pKa = 10.93DD225 pKa = 2.84KK226 pKa = 10.3VTLLRR231 pKa = 11.84SGNANGTIKK240 pKa = 10.19EE241 pKa = 4.03AKK243 pKa = 8.93LWHH246 pKa = 6.09GMNKK250 pKa = 9.33NLQYY254 pKa = 11.48DD255 pKa = 3.82DD256 pKa = 5.66DD257 pKa = 4.59EE258 pKa = 6.84SGDD261 pKa = 4.01KK262 pKa = 10.86EE263 pKa = 4.46STSYY267 pKa = 11.12TSVDD271 pKa = 3.4SKK273 pKa = 11.23IGMGDD278 pKa = 3.74YY279 pKa = 10.75FVMDD283 pKa = 3.84MFYY286 pKa = 10.52PGAGGAAADD295 pKa = 4.34IILVQANSTLYY306 pKa = 9.1WHH308 pKa = 7.05EE309 pKa = 4.08KK310 pKa = 9.32
MM1 pKa = 7.78AYY3 pKa = 9.79RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 10.07SYY8 pKa = 10.46SSRR11 pKa = 11.84ARR13 pKa = 11.84RR14 pKa = 11.84PIGRR18 pKa = 11.84SAKK21 pKa = 10.01RR22 pKa = 11.84GGARR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 5.34PTKK32 pKa = 10.21KK33 pKa = 8.56RR34 pKa = 11.84TYY36 pKa = 8.62RR37 pKa = 11.84KK38 pKa = 9.1NRR40 pKa = 11.84AMSTRR45 pKa = 11.84TILNKK50 pKa = 9.28TSRR53 pKa = 11.84KK54 pKa = 8.91KK55 pKa = 10.61RR56 pKa = 11.84NGMLTWANTSTSGTSIPNVATNYY79 pKa = 10.18FLTGNLGGRR88 pKa = 11.84SIWCATAQDD97 pKa = 4.27LTVQDD102 pKa = 4.27GSFGTVALAAARR114 pKa = 11.84TSTSCYY120 pKa = 8.69MKK122 pKa = 10.59GLSEE126 pKa = 5.06HH127 pKa = 6.62IRR129 pKa = 11.84IQTSTGLPWFWRR141 pKa = 11.84RR142 pKa = 11.84ICFTLKK148 pKa = 10.67GDD150 pKa = 3.82DD151 pKa = 3.83LFRR154 pKa = 11.84FSTSDD159 pKa = 3.31TPTSTSNRR167 pKa = 11.84YY168 pKa = 9.6LDD170 pKa = 3.69TTGGMQRR177 pKa = 11.84LFFNEE182 pKa = 3.79LNNAQSNTINNHH194 pKa = 5.19DD195 pKa = 4.5AIIFKK200 pKa = 9.9GAKK203 pKa = 9.23GIDD206 pKa = 3.34WIDD209 pKa = 4.2YY210 pKa = 9.29IVAPIDD216 pKa = 3.37TSRR219 pKa = 11.84INLKK223 pKa = 9.81YY224 pKa = 10.93DD225 pKa = 2.84KK226 pKa = 10.3VTLLRR231 pKa = 11.84SGNANGTIKK240 pKa = 10.19EE241 pKa = 4.03AKK243 pKa = 8.93LWHH246 pKa = 6.09GMNKK250 pKa = 9.33NLQYY254 pKa = 11.48DD255 pKa = 3.82DD256 pKa = 5.66DD257 pKa = 4.59EE258 pKa = 6.84SGDD261 pKa = 4.01KK262 pKa = 10.86EE263 pKa = 4.46STSYY267 pKa = 11.12TSVDD271 pKa = 3.4SKK273 pKa = 11.23IGMGDD278 pKa = 3.74YY279 pKa = 10.75FVMDD283 pKa = 3.84MFYY286 pKa = 10.52PGAGGAAADD295 pKa = 4.34IILVQANSTLYY306 pKa = 9.1WHH308 pKa = 7.05EE309 pKa = 4.08KK310 pKa = 9.32
Molecular weight: 34.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
645 |
310 |
335 |
322.5 |
36.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.372 ± 0.433 | 1.395 ± 0.293 |
6.977 ± 0.582 | 3.721 ± 1.225 |
4.341 ± 0.544 | 8.527 ± 0.096 |
2.326 ± 0.711 | 6.202 ± 0.05 |
5.736 ± 0.491 | 6.977 ± 0.36 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.326 ± 0.396 | 5.116 ± 0.917 |
3.256 ± 0.685 | 2.481 ± 0.153 |
7.287 ± 0.755 | 7.287 ± 1.198 |
6.822 ± 2.403 | 4.186 ± 1.102 |
2.636 ± 0.259 | 4.031 ± 0.333 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
