
Circovirus-like genome DCCV-5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.84
Get precalculated fractions of proteins
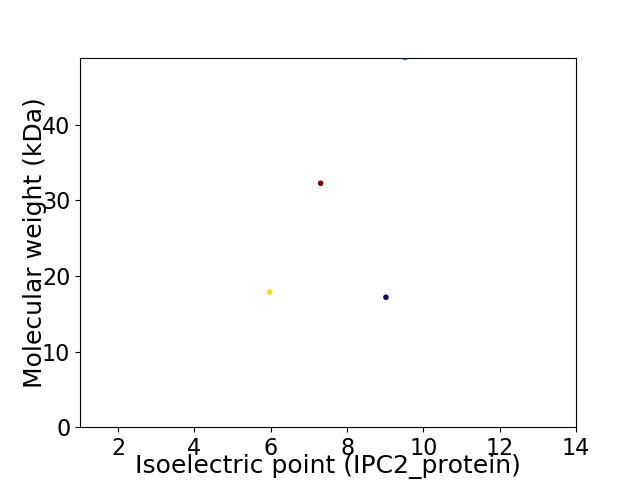
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
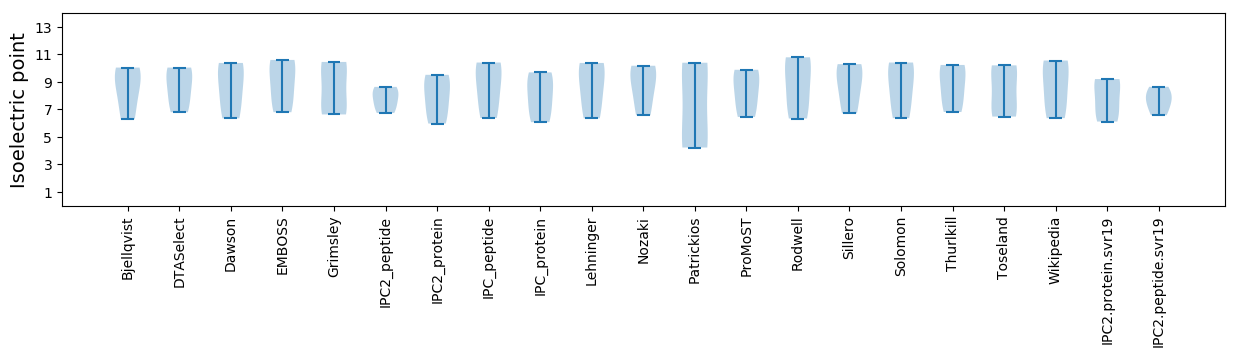
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A190WHA9|A0A190WHA9_9CIRC Uncharacterized protein OS=Circovirus-like genome DCCV-5 OX=1788445 PE=4 SV=1
MM1 pKa = 6.7TTQLLTPGSTHH12 pKa = 6.42SGHH15 pKa = 6.84SIALPVGRR23 pKa = 11.84SPLVPEE29 pKa = 4.06HH30 pKa = 5.54LTYY33 pKa = 10.62RR34 pKa = 11.84DD35 pKa = 3.73TPSTLIRR42 pKa = 11.84SPSPPLDD49 pKa = 4.47DD50 pKa = 4.53YY51 pKa = 11.74LWDD54 pKa = 4.09VTWSPPVGPDD64 pKa = 3.43PNPEE68 pKa = 3.67LTALRR73 pKa = 11.84MAAFARR79 pKa = 11.84EE80 pKa = 4.16VSPPPMRR87 pKa = 11.84TPGTGRR93 pKa = 11.84SSVGRR98 pKa = 11.84TPVPWLRR105 pKa = 11.84RR106 pKa = 11.84ASSTRR111 pKa = 11.84FPLIYY116 pKa = 10.32TSDD119 pKa = 3.4TSEE122 pKa = 4.03TSTGSIEE129 pKa = 4.26RR130 pKa = 11.84FSRR133 pKa = 11.84PWSPSPLPAGVGSWEE148 pKa = 4.07EE149 pKa = 3.64LDD151 pKa = 3.88LVNLRR156 pKa = 11.84EE157 pKa = 4.26CDD159 pKa = 3.36QPSRR163 pKa = 3.86
MM1 pKa = 6.7TTQLLTPGSTHH12 pKa = 6.42SGHH15 pKa = 6.84SIALPVGRR23 pKa = 11.84SPLVPEE29 pKa = 4.06HH30 pKa = 5.54LTYY33 pKa = 10.62RR34 pKa = 11.84DD35 pKa = 3.73TPSTLIRR42 pKa = 11.84SPSPPLDD49 pKa = 4.47DD50 pKa = 4.53YY51 pKa = 11.74LWDD54 pKa = 4.09VTWSPPVGPDD64 pKa = 3.43PNPEE68 pKa = 3.67LTALRR73 pKa = 11.84MAAFARR79 pKa = 11.84EE80 pKa = 4.16VSPPPMRR87 pKa = 11.84TPGTGRR93 pKa = 11.84SSVGRR98 pKa = 11.84TPVPWLRR105 pKa = 11.84RR106 pKa = 11.84ASSTRR111 pKa = 11.84FPLIYY116 pKa = 10.32TSDD119 pKa = 3.4TSEE122 pKa = 4.03TSTGSIEE129 pKa = 4.26RR130 pKa = 11.84FSRR133 pKa = 11.84PWSPSPLPAGVGSWEE148 pKa = 4.07EE149 pKa = 3.64LDD151 pKa = 3.88LVNLRR156 pKa = 11.84EE157 pKa = 4.26CDD159 pKa = 3.36QPSRR163 pKa = 3.86
Molecular weight: 17.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A190WHJ3|A0A190WHJ3_9CIRC Uncharacterized protein OS=Circovirus-like genome DCCV-5 OX=1788445 PE=4 SV=1
MM1 pKa = 7.42AAYY4 pKa = 9.41AARR7 pKa = 11.84IGYY10 pKa = 9.3GLARR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 9.08AVPAIRR22 pKa = 11.84RR23 pKa = 11.84SIPAIQRR30 pKa = 11.84FARR33 pKa = 11.84RR34 pKa = 11.84SFVPRR39 pKa = 11.84QNGLGIPLRR48 pKa = 11.84RR49 pKa = 11.84IAPSRR54 pKa = 11.84AANFNQIRR62 pKa = 11.84RR63 pKa = 11.84MGSTAFPRR71 pKa = 11.84TPTSNPIPSTSRR83 pKa = 11.84AINRR87 pKa = 11.84IARR90 pKa = 11.84GATAAAATGGALKK103 pKa = 10.62ALSNAKK109 pKa = 9.32KK110 pKa = 10.38RR111 pKa = 11.84KK112 pKa = 9.87AEE114 pKa = 4.03EE115 pKa = 3.49QNKK118 pKa = 9.35RR119 pKa = 11.84VKK121 pKa = 10.57KK122 pKa = 10.52DD123 pKa = 3.08IEE125 pKa = 3.99QNAASGLGDD134 pKa = 4.04DD135 pKa = 4.75VKK137 pKa = 10.6TMDD140 pKa = 4.53SFTHH144 pKa = 6.63HH145 pKa = 6.88SGNDD149 pKa = 3.1SWLKK153 pKa = 9.98AHH155 pKa = 6.83LVSGKK160 pKa = 8.44HH161 pKa = 4.31TVKK164 pKa = 10.84NLAFSNNLLGTQGRR178 pKa = 11.84ARR180 pKa = 11.84WFTVTTTQTPDD191 pKa = 3.05DD192 pKa = 4.24YY193 pKa = 10.99RR194 pKa = 11.84TLFQQTTEE202 pKa = 4.2NYY204 pKa = 9.58RR205 pKa = 11.84STSTASATPSWVRR218 pKa = 11.84GDD220 pKa = 4.26PLLQSGYY227 pKa = 10.37LNQKK231 pKa = 9.38FVYY234 pKa = 9.54EE235 pKa = 4.11KK236 pKa = 10.97LKK238 pKa = 11.1ANYY241 pKa = 9.04IFKK244 pKa = 10.33NQGSSPVHH252 pKa = 5.12MEE254 pKa = 3.67YY255 pKa = 10.61FIITPKK261 pKa = 9.54DD262 pKa = 3.35TNEE265 pKa = 4.68IARR268 pKa = 11.84NWQNDD273 pKa = 3.75YY274 pKa = 11.8VNGLSDD280 pKa = 3.67STAEE284 pKa = 4.19LAVKK288 pKa = 10.47ASISSGVLVTPCEE301 pKa = 4.11YY302 pKa = 10.7KK303 pKa = 10.27IGSSPRR309 pKa = 11.84FNKK312 pKa = 9.06YY313 pKa = 7.22WKK315 pKa = 9.36ILYY318 pKa = 7.86SHH320 pKa = 7.5KK321 pKa = 10.72VRR323 pKa = 11.84MAEE326 pKa = 4.08GAEE329 pKa = 4.14HH330 pKa = 6.79EE331 pKa = 5.51FNFTDD336 pKa = 3.27NANRR340 pKa = 11.84EE341 pKa = 4.28IQWNGFPVNGGQLKK355 pKa = 10.57GMTKK359 pKa = 9.75YY360 pKa = 10.62FIVKK364 pKa = 9.72IYY366 pKa = 10.95GDD368 pKa = 3.82IGDD371 pKa = 4.24SSNLLGTIGTITTAPAKK388 pKa = 10.24IIGILRR394 pKa = 11.84HH395 pKa = 5.55TEE397 pKa = 4.13TVRR400 pKa = 11.84HH401 pKa = 4.52VHH403 pKa = 5.59RR404 pKa = 11.84TPSLFLDD411 pKa = 5.3FSADD415 pKa = 3.47LATNAAALYY424 pKa = 9.38EE425 pKa = 4.3KK426 pKa = 10.72DD427 pKa = 3.32GGTGKK432 pKa = 10.24VDD434 pKa = 3.41NVLLEE439 pKa = 4.16VQPP442 pKa = 4.25
MM1 pKa = 7.42AAYY4 pKa = 9.41AARR7 pKa = 11.84IGYY10 pKa = 9.3GLARR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 9.08AVPAIRR22 pKa = 11.84RR23 pKa = 11.84SIPAIQRR30 pKa = 11.84FARR33 pKa = 11.84RR34 pKa = 11.84SFVPRR39 pKa = 11.84QNGLGIPLRR48 pKa = 11.84RR49 pKa = 11.84IAPSRR54 pKa = 11.84AANFNQIRR62 pKa = 11.84RR63 pKa = 11.84MGSTAFPRR71 pKa = 11.84TPTSNPIPSTSRR83 pKa = 11.84AINRR87 pKa = 11.84IARR90 pKa = 11.84GATAAAATGGALKK103 pKa = 10.62ALSNAKK109 pKa = 9.32KK110 pKa = 10.38RR111 pKa = 11.84KK112 pKa = 9.87AEE114 pKa = 4.03EE115 pKa = 3.49QNKK118 pKa = 9.35RR119 pKa = 11.84VKK121 pKa = 10.57KK122 pKa = 10.52DD123 pKa = 3.08IEE125 pKa = 3.99QNAASGLGDD134 pKa = 4.04DD135 pKa = 4.75VKK137 pKa = 10.6TMDD140 pKa = 4.53SFTHH144 pKa = 6.63HH145 pKa = 6.88SGNDD149 pKa = 3.1SWLKK153 pKa = 9.98AHH155 pKa = 6.83LVSGKK160 pKa = 8.44HH161 pKa = 4.31TVKK164 pKa = 10.84NLAFSNNLLGTQGRR178 pKa = 11.84ARR180 pKa = 11.84WFTVTTTQTPDD191 pKa = 3.05DD192 pKa = 4.24YY193 pKa = 10.99RR194 pKa = 11.84TLFQQTTEE202 pKa = 4.2NYY204 pKa = 9.58RR205 pKa = 11.84STSTASATPSWVRR218 pKa = 11.84GDD220 pKa = 4.26PLLQSGYY227 pKa = 10.37LNQKK231 pKa = 9.38FVYY234 pKa = 9.54EE235 pKa = 4.11KK236 pKa = 10.97LKK238 pKa = 11.1ANYY241 pKa = 9.04IFKK244 pKa = 10.33NQGSSPVHH252 pKa = 5.12MEE254 pKa = 3.67YY255 pKa = 10.61FIITPKK261 pKa = 9.54DD262 pKa = 3.35TNEE265 pKa = 4.68IARR268 pKa = 11.84NWQNDD273 pKa = 3.75YY274 pKa = 11.8VNGLSDD280 pKa = 3.67STAEE284 pKa = 4.19LAVKK288 pKa = 10.47ASISSGVLVTPCEE301 pKa = 4.11YY302 pKa = 10.7KK303 pKa = 10.27IGSSPRR309 pKa = 11.84FNKK312 pKa = 9.06YY313 pKa = 7.22WKK315 pKa = 9.36ILYY318 pKa = 7.86SHH320 pKa = 7.5KK321 pKa = 10.72VRR323 pKa = 11.84MAEE326 pKa = 4.08GAEE329 pKa = 4.14HH330 pKa = 6.79EE331 pKa = 5.51FNFTDD336 pKa = 3.27NANRR340 pKa = 11.84EE341 pKa = 4.28IQWNGFPVNGGQLKK355 pKa = 10.57GMTKK359 pKa = 9.75YY360 pKa = 10.62FIVKK364 pKa = 9.72IYY366 pKa = 10.95GDD368 pKa = 3.82IGDD371 pKa = 4.24SSNLLGTIGTITTAPAKK388 pKa = 10.24IIGILRR394 pKa = 11.84HH395 pKa = 5.55TEE397 pKa = 4.13TVRR400 pKa = 11.84HH401 pKa = 4.52VHH403 pKa = 5.59RR404 pKa = 11.84TPSLFLDD411 pKa = 5.3FSADD415 pKa = 3.47LATNAAALYY424 pKa = 9.38EE425 pKa = 4.3KK426 pKa = 10.72DD427 pKa = 3.32GGTGKK432 pKa = 10.24VDD434 pKa = 3.41NVLLEE439 pKa = 4.16VQPP442 pKa = 4.25
Molecular weight: 48.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1034 |
148 |
442 |
258.5 |
29.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.544 ± 1.338 | 0.774 ± 0.304 |
4.739 ± 0.41 | 4.932 ± 0.64 |
3.385 ± 0.425 | 7.06 ± 0.441 |
2.611 ± 0.454 | 5.222 ± 0.888 |
4.739 ± 1.102 | 8.414 ± 0.892 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.451 ± 0.311 | 4.739 ± 1.12 |
7.06 ± 1.923 | 3.675 ± 1.004 |
8.027 ± 0.404 | 8.027 ± 1.221 |
7.35 ± 0.959 | 4.545 ± 0.544 |
2.224 ± 0.398 | 3.482 ± 0.557 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
