
Sanxia sobemo-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.7
Get precalculated fractions of proteins
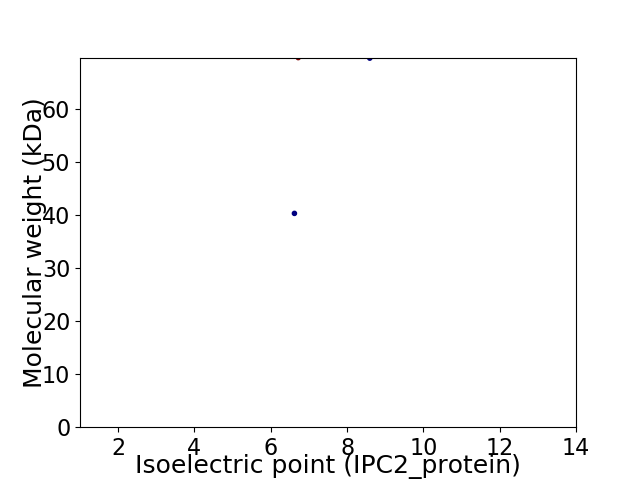
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEW7|A0A1L3KEW7_9VIRU C2H2-type domain-containing protein OS=Sanxia sobemo-like virus 5 OX=1923384 PE=4 SV=1
MM1 pKa = 7.92SYY3 pKa = 9.73EE4 pKa = 3.77HH5 pKa = 6.57YY6 pKa = 10.62EE7 pKa = 4.38RR8 pKa = 11.84VVQDD12 pKa = 4.56LDD14 pKa = 3.56WTSSPGYY21 pKa = 9.33PYY23 pKa = 11.21LLNAPTNGSFFQVQNGIPSEE43 pKa = 4.14EE44 pKa = 4.05AKK46 pKa = 10.71HH47 pKa = 6.2RR48 pKa = 11.84VWQLLTQRR56 pKa = 11.84LLDD59 pKa = 4.02RR60 pKa = 11.84SCDD63 pKa = 4.05PIRR66 pKa = 11.84LFIKK70 pKa = 10.31PEE72 pKa = 3.66PHH74 pKa = 6.13KK75 pKa = 10.48VAKK78 pKa = 10.36IEE80 pKa = 3.95TGRR83 pKa = 11.84FRR85 pKa = 11.84LISAVSVVDD94 pKa = 4.22QIIDD98 pKa = 3.43SMLFGEE104 pKa = 4.67MNKK107 pKa = 10.13NMVRR111 pKa = 11.84HH112 pKa = 5.91CLEE115 pKa = 4.2IPSKK119 pKa = 10.57FGWSPYY125 pKa = 8.31VGGWKK130 pKa = 9.76VVPQKK135 pKa = 11.1GLAIDD140 pKa = 3.6KK141 pKa = 10.77SSWDD145 pKa = 2.98WTMQGWLFQIILEE158 pKa = 4.18LRR160 pKa = 11.84SRR162 pKa = 11.84LCLTVGKK169 pKa = 9.73RR170 pKa = 11.84HH171 pKa = 6.49KK172 pKa = 10.06LWQEE176 pKa = 3.67LASWRR181 pKa = 11.84YY182 pKa = 8.13EE183 pKa = 3.77ALFGSPEE190 pKa = 4.31FILSNGLVLQQSQPGVMKK208 pKa = 10.24SGCVNTIADD217 pKa = 3.59NSIAQVVLHH226 pKa = 6.26LRR228 pKa = 11.84VCLEE232 pKa = 3.71MGIPAGWIWSMGDD245 pKa = 3.34DD246 pKa = 3.73TLQEE250 pKa = 4.1PLRR253 pKa = 11.84DD254 pKa = 3.59EE255 pKa = 4.74GSYY258 pKa = 10.39LASLSRR264 pKa = 11.84YY265 pKa = 9.6CIVKK269 pKa = 10.33EE270 pKa = 4.06STHH273 pKa = 7.01LIEE276 pKa = 4.94FAGHH280 pKa = 5.49QFRR283 pKa = 11.84KK284 pKa = 10.45DD285 pKa = 3.65SIEE288 pKa = 3.51PSYY291 pKa = 10.13FQKK294 pKa = 10.78HH295 pKa = 5.63CFNLLHH301 pKa = 7.06ADD303 pKa = 3.61SDD305 pKa = 4.51VVADD309 pKa = 4.41LAVSYY314 pKa = 11.27ALLYY318 pKa = 10.5HH319 pKa = 6.6KK320 pKa = 10.35SKK322 pKa = 10.73KK323 pKa = 9.56RR324 pKa = 11.84DD325 pKa = 4.25FIHH328 pKa = 6.68HH329 pKa = 6.67VIRR332 pKa = 11.84EE333 pKa = 4.19LGGIPASPRR342 pKa = 11.84MIAQIYY348 pKa = 8.79DD349 pKa = 3.89GPWW352 pKa = 2.67
MM1 pKa = 7.92SYY3 pKa = 9.73EE4 pKa = 3.77HH5 pKa = 6.57YY6 pKa = 10.62EE7 pKa = 4.38RR8 pKa = 11.84VVQDD12 pKa = 4.56LDD14 pKa = 3.56WTSSPGYY21 pKa = 9.33PYY23 pKa = 11.21LLNAPTNGSFFQVQNGIPSEE43 pKa = 4.14EE44 pKa = 4.05AKK46 pKa = 10.71HH47 pKa = 6.2RR48 pKa = 11.84VWQLLTQRR56 pKa = 11.84LLDD59 pKa = 4.02RR60 pKa = 11.84SCDD63 pKa = 4.05PIRR66 pKa = 11.84LFIKK70 pKa = 10.31PEE72 pKa = 3.66PHH74 pKa = 6.13KK75 pKa = 10.48VAKK78 pKa = 10.36IEE80 pKa = 3.95TGRR83 pKa = 11.84FRR85 pKa = 11.84LISAVSVVDD94 pKa = 4.22QIIDD98 pKa = 3.43SMLFGEE104 pKa = 4.67MNKK107 pKa = 10.13NMVRR111 pKa = 11.84HH112 pKa = 5.91CLEE115 pKa = 4.2IPSKK119 pKa = 10.57FGWSPYY125 pKa = 8.31VGGWKK130 pKa = 9.76VVPQKK135 pKa = 11.1GLAIDD140 pKa = 3.6KK141 pKa = 10.77SSWDD145 pKa = 2.98WTMQGWLFQIILEE158 pKa = 4.18LRR160 pKa = 11.84SRR162 pKa = 11.84LCLTVGKK169 pKa = 9.73RR170 pKa = 11.84HH171 pKa = 6.49KK172 pKa = 10.06LWQEE176 pKa = 3.67LASWRR181 pKa = 11.84YY182 pKa = 8.13EE183 pKa = 3.77ALFGSPEE190 pKa = 4.31FILSNGLVLQQSQPGVMKK208 pKa = 10.24SGCVNTIADD217 pKa = 3.59NSIAQVVLHH226 pKa = 6.26LRR228 pKa = 11.84VCLEE232 pKa = 3.71MGIPAGWIWSMGDD245 pKa = 3.34DD246 pKa = 3.73TLQEE250 pKa = 4.1PLRR253 pKa = 11.84DD254 pKa = 3.59EE255 pKa = 4.74GSYY258 pKa = 10.39LASLSRR264 pKa = 11.84YY265 pKa = 9.6CIVKK269 pKa = 10.33EE270 pKa = 4.06STHH273 pKa = 7.01LIEE276 pKa = 4.94FAGHH280 pKa = 5.49QFRR283 pKa = 11.84KK284 pKa = 10.45DD285 pKa = 3.65SIEE288 pKa = 3.51PSYY291 pKa = 10.13FQKK294 pKa = 10.78HH295 pKa = 5.63CFNLLHH301 pKa = 7.06ADD303 pKa = 3.61SDD305 pKa = 4.51VVADD309 pKa = 4.41LAVSYY314 pKa = 11.27ALLYY318 pKa = 10.5HH319 pKa = 6.6KK320 pKa = 10.35SKK322 pKa = 10.73KK323 pKa = 9.56RR324 pKa = 11.84DD325 pKa = 4.25FIHH328 pKa = 6.68HH329 pKa = 6.67VIRR332 pKa = 11.84EE333 pKa = 4.19LGGIPASPRR342 pKa = 11.84MIAQIYY348 pKa = 8.79DD349 pKa = 3.89GPWW352 pKa = 2.67
Molecular weight: 40.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEW7|A0A1L3KEW7_9VIRU C2H2-type domain-containing protein OS=Sanxia sobemo-like virus 5 OX=1923384 PE=4 SV=1
MM1 pKa = 7.32VFGDD5 pKa = 3.93LTSCIRR11 pKa = 11.84MGYY14 pKa = 7.7NHH16 pKa = 6.51SFIRR20 pKa = 11.84RR21 pKa = 11.84FVVSAVATQLIIVSYY36 pKa = 10.49GFVSRR41 pKa = 11.84LAQWVVLVLSHH52 pKa = 7.32LLGPLFLGAVEE63 pKa = 4.7GIEE66 pKa = 3.98VGGIKK71 pKa = 9.88PYY73 pKa = 10.72KK74 pKa = 9.37LWLEE78 pKa = 4.38VGVEE82 pKa = 4.28PPRR85 pKa = 11.84PPAYY89 pKa = 9.83QRR91 pKa = 11.84LVDD94 pKa = 6.27GICEE98 pKa = 4.22WILNHH103 pKa = 5.53EE104 pKa = 4.56QEE106 pKa = 4.38TSMMLGVFACLVMLRR121 pKa = 11.84LLFKK125 pKa = 10.99LPVRR129 pKa = 11.84RR130 pKa = 11.84MSYY133 pKa = 9.87KK134 pKa = 10.06LRR136 pKa = 11.84GIRR139 pKa = 11.84FEE141 pKa = 4.23SVKK144 pKa = 10.64PGSEE148 pKa = 3.61LLAGVKK154 pKa = 10.07EE155 pKa = 4.27PAFQPRR161 pKa = 11.84IMQPGLFVNSFQGYY175 pKa = 8.85GIRR178 pKa = 11.84LGKK181 pKa = 9.7FLVMPRR187 pKa = 11.84HH188 pKa = 6.17VYY190 pKa = 9.45DD191 pKa = 3.44ACEE194 pKa = 4.19GQVMMRR200 pKa = 11.84GPKK203 pKa = 9.61SSYY206 pKa = 10.17IIANAPIQSRR216 pKa = 11.84VVRR219 pKa = 11.84DD220 pKa = 3.31LVYY223 pKa = 10.4IPLAEE228 pKa = 4.78KK229 pKa = 10.71VFSDD233 pKa = 4.55CGIPSATMARR243 pKa = 11.84RR244 pKa = 11.84LAKK247 pKa = 10.2GALVTCIGEE256 pKa = 4.13QGASSGIIRR265 pKa = 11.84PLSIMGMLSYY275 pKa = 10.22TGSTVPGMSGAAYY288 pKa = 9.17IVNSMSRR295 pKa = 11.84KK296 pKa = 7.77VHH298 pKa = 6.5GIHH301 pKa = 6.64HH302 pKa = 6.07GVVGEE307 pKa = 4.22NNVGTSSLIVAKK319 pKa = 9.22EE320 pKa = 3.67LKK322 pKa = 10.33ALEE325 pKa = 3.94GRR327 pKa = 11.84MFGEE331 pKa = 4.23SPLGNEE337 pKa = 4.63VEE339 pKa = 4.25EE340 pKa = 4.16QLADD344 pKa = 3.52RR345 pKa = 11.84RR346 pKa = 11.84GGYY349 pKa = 8.85KK350 pKa = 9.92AWAEE354 pKa = 4.0DD355 pKa = 3.88DD356 pKa = 3.87LDD358 pKa = 4.5EE359 pKa = 4.72VVLDD363 pKa = 3.62AWEE366 pKa = 4.95ADD368 pKa = 3.92DD369 pKa = 6.56DD370 pKa = 4.02DD371 pKa = 4.04WAYY374 pKa = 11.24DD375 pKa = 4.09DD376 pKa = 5.89DD377 pKa = 6.54AMDD380 pKa = 3.8YY381 pKa = 11.22EE382 pKa = 4.64EE383 pKa = 5.01DD384 pKa = 4.14VVWSRR389 pKa = 11.84EE390 pKa = 4.17SEE392 pKa = 5.32DD393 pKa = 3.32EE394 pKa = 4.36DD395 pKa = 4.04FWDD398 pKa = 4.02EE399 pKa = 5.12GPTRR403 pKa = 11.84WEE405 pKa = 3.73SRR407 pKa = 11.84KK408 pKa = 9.55RR409 pKa = 11.84RR410 pKa = 11.84PRR412 pKa = 11.84KK413 pKa = 8.55KK414 pKa = 8.56KK415 pKa = 8.92AVRR418 pKa = 11.84RR419 pKa = 11.84VVGLGHH425 pKa = 6.68AASRR429 pKa = 11.84PEE431 pKa = 3.67IEE433 pKa = 3.85ISGKK437 pKa = 9.94VKK439 pKa = 10.27HH440 pKa = 5.81QSGPQAQSKK449 pKa = 10.49ASPEE453 pKa = 3.77KK454 pKa = 10.25AAPPEE459 pKa = 4.06KK460 pKa = 9.79TVRR463 pKa = 11.84QAFNEE468 pKa = 3.95LEE470 pKa = 3.98ARR472 pKa = 11.84VSKK475 pKa = 10.95LEE477 pKa = 3.76KK478 pKa = 9.95RR479 pKa = 11.84VSSLEE484 pKa = 3.46ARR486 pKa = 11.84LPDD489 pKa = 3.61RR490 pKa = 11.84KK491 pKa = 9.96EE492 pKa = 3.76VAQKK496 pKa = 10.4VDD498 pKa = 3.31KK499 pKa = 10.83AVSAVRR505 pKa = 11.84TRR507 pKa = 11.84PEE509 pKa = 3.9PKK511 pKa = 9.63RR512 pKa = 11.84PPGSVVCMEE521 pKa = 4.69CKK523 pKa = 9.93RR524 pKa = 11.84RR525 pKa = 11.84FPHH528 pKa = 5.63VRR530 pKa = 11.84ALVAHH535 pKa = 6.78NDD537 pKa = 3.12EE538 pKa = 4.04KK539 pKa = 11.31HH540 pKa = 5.07RR541 pKa = 11.84VKK543 pKa = 11.04GEE545 pKa = 3.94SALKK549 pKa = 9.77EE550 pKa = 3.97DD551 pKa = 3.62SKK553 pKa = 11.64KK554 pKa = 10.55VVKK557 pKa = 7.02TTRR560 pKa = 11.84GRR562 pKa = 11.84FLGNSPKK569 pKa = 10.66NPGDD573 pKa = 3.68GPRR576 pKa = 11.84GSSKK580 pKa = 10.67LSPRR584 pKa = 11.84SSQFRR589 pKa = 11.84SILEE593 pKa = 4.35SQSKK597 pKa = 9.82MSDD600 pKa = 3.34CLCRR604 pKa = 11.84LEE606 pKa = 5.47KK607 pKa = 11.04SFDD610 pKa = 3.56LLARR614 pKa = 11.84AMAGPSSEE622 pKa = 3.88PRR624 pKa = 11.84RR625 pKa = 11.84NN626 pKa = 3.18
MM1 pKa = 7.32VFGDD5 pKa = 3.93LTSCIRR11 pKa = 11.84MGYY14 pKa = 7.7NHH16 pKa = 6.51SFIRR20 pKa = 11.84RR21 pKa = 11.84FVVSAVATQLIIVSYY36 pKa = 10.49GFVSRR41 pKa = 11.84LAQWVVLVLSHH52 pKa = 7.32LLGPLFLGAVEE63 pKa = 4.7GIEE66 pKa = 3.98VGGIKK71 pKa = 9.88PYY73 pKa = 10.72KK74 pKa = 9.37LWLEE78 pKa = 4.38VGVEE82 pKa = 4.28PPRR85 pKa = 11.84PPAYY89 pKa = 9.83QRR91 pKa = 11.84LVDD94 pKa = 6.27GICEE98 pKa = 4.22WILNHH103 pKa = 5.53EE104 pKa = 4.56QEE106 pKa = 4.38TSMMLGVFACLVMLRR121 pKa = 11.84LLFKK125 pKa = 10.99LPVRR129 pKa = 11.84RR130 pKa = 11.84MSYY133 pKa = 9.87KK134 pKa = 10.06LRR136 pKa = 11.84GIRR139 pKa = 11.84FEE141 pKa = 4.23SVKK144 pKa = 10.64PGSEE148 pKa = 3.61LLAGVKK154 pKa = 10.07EE155 pKa = 4.27PAFQPRR161 pKa = 11.84IMQPGLFVNSFQGYY175 pKa = 8.85GIRR178 pKa = 11.84LGKK181 pKa = 9.7FLVMPRR187 pKa = 11.84HH188 pKa = 6.17VYY190 pKa = 9.45DD191 pKa = 3.44ACEE194 pKa = 4.19GQVMMRR200 pKa = 11.84GPKK203 pKa = 9.61SSYY206 pKa = 10.17IIANAPIQSRR216 pKa = 11.84VVRR219 pKa = 11.84DD220 pKa = 3.31LVYY223 pKa = 10.4IPLAEE228 pKa = 4.78KK229 pKa = 10.71VFSDD233 pKa = 4.55CGIPSATMARR243 pKa = 11.84RR244 pKa = 11.84LAKK247 pKa = 10.2GALVTCIGEE256 pKa = 4.13QGASSGIIRR265 pKa = 11.84PLSIMGMLSYY275 pKa = 10.22TGSTVPGMSGAAYY288 pKa = 9.17IVNSMSRR295 pKa = 11.84KK296 pKa = 7.77VHH298 pKa = 6.5GIHH301 pKa = 6.64HH302 pKa = 6.07GVVGEE307 pKa = 4.22NNVGTSSLIVAKK319 pKa = 9.22EE320 pKa = 3.67LKK322 pKa = 10.33ALEE325 pKa = 3.94GRR327 pKa = 11.84MFGEE331 pKa = 4.23SPLGNEE337 pKa = 4.63VEE339 pKa = 4.25EE340 pKa = 4.16QLADD344 pKa = 3.52RR345 pKa = 11.84RR346 pKa = 11.84GGYY349 pKa = 8.85KK350 pKa = 9.92AWAEE354 pKa = 4.0DD355 pKa = 3.88DD356 pKa = 3.87LDD358 pKa = 4.5EE359 pKa = 4.72VVLDD363 pKa = 3.62AWEE366 pKa = 4.95ADD368 pKa = 3.92DD369 pKa = 6.56DD370 pKa = 4.02DD371 pKa = 4.04WAYY374 pKa = 11.24DD375 pKa = 4.09DD376 pKa = 5.89DD377 pKa = 6.54AMDD380 pKa = 3.8YY381 pKa = 11.22EE382 pKa = 4.64EE383 pKa = 5.01DD384 pKa = 4.14VVWSRR389 pKa = 11.84EE390 pKa = 4.17SEE392 pKa = 5.32DD393 pKa = 3.32EE394 pKa = 4.36DD395 pKa = 4.04FWDD398 pKa = 4.02EE399 pKa = 5.12GPTRR403 pKa = 11.84WEE405 pKa = 3.73SRR407 pKa = 11.84KK408 pKa = 9.55RR409 pKa = 11.84RR410 pKa = 11.84PRR412 pKa = 11.84KK413 pKa = 8.55KK414 pKa = 8.56KK415 pKa = 8.92AVRR418 pKa = 11.84RR419 pKa = 11.84VVGLGHH425 pKa = 6.68AASRR429 pKa = 11.84PEE431 pKa = 3.67IEE433 pKa = 3.85ISGKK437 pKa = 9.94VKK439 pKa = 10.27HH440 pKa = 5.81QSGPQAQSKK449 pKa = 10.49ASPEE453 pKa = 3.77KK454 pKa = 10.25AAPPEE459 pKa = 4.06KK460 pKa = 9.79TVRR463 pKa = 11.84QAFNEE468 pKa = 3.95LEE470 pKa = 3.98ARR472 pKa = 11.84VSKK475 pKa = 10.95LEE477 pKa = 3.76KK478 pKa = 9.95RR479 pKa = 11.84VSSLEE484 pKa = 3.46ARR486 pKa = 11.84LPDD489 pKa = 3.61RR490 pKa = 11.84KK491 pKa = 9.96EE492 pKa = 3.76VAQKK496 pKa = 10.4VDD498 pKa = 3.31KK499 pKa = 10.83AVSAVRR505 pKa = 11.84TRR507 pKa = 11.84PEE509 pKa = 3.9PKK511 pKa = 9.63RR512 pKa = 11.84PPGSVVCMEE521 pKa = 4.69CKK523 pKa = 9.93RR524 pKa = 11.84RR525 pKa = 11.84FPHH528 pKa = 5.63VRR530 pKa = 11.84ALVAHH535 pKa = 6.78NDD537 pKa = 3.12EE538 pKa = 4.04KK539 pKa = 11.31HH540 pKa = 5.07RR541 pKa = 11.84VKK543 pKa = 11.04GEE545 pKa = 3.94SALKK549 pKa = 9.77EE550 pKa = 3.97DD551 pKa = 3.62SKK553 pKa = 11.64KK554 pKa = 10.55VVKK557 pKa = 7.02TTRR560 pKa = 11.84GRR562 pKa = 11.84FLGNSPKK569 pKa = 10.66NPGDD573 pKa = 3.68GPRR576 pKa = 11.84GSSKK580 pKa = 10.67LSPRR584 pKa = 11.84SSQFRR589 pKa = 11.84SILEE593 pKa = 4.35SQSKK597 pKa = 9.82MSDD600 pKa = 3.34CLCRR604 pKa = 11.84LEE606 pKa = 5.47KK607 pKa = 11.04SFDD610 pKa = 3.56LLARR614 pKa = 11.84AMAGPSSEE622 pKa = 3.88PRR624 pKa = 11.84RR625 pKa = 11.84NN626 pKa = 3.18
Molecular weight: 69.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
978 |
352 |
626 |
489.0 |
54.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.646 ± 0.933 | 1.738 ± 0.152 |
4.806 ± 0.187 | 6.748 ± 0.822 |
3.476 ± 0.305 | 7.566 ± 0.628 |
2.556 ± 0.692 | 5.112 ± 1.038 |
5.93 ± 0.497 | 9.1 ± 0.859 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.965 ± 0.249 | 2.249 ± 0.187 |
5.726 ± 0.373 | 3.681 ± 0.872 |
7.464 ± 1.258 | 8.793 ± 0.008 |
2.249 ± 0.187 | 8.384 ± 0.953 |
2.147 ± 0.768 | 2.658 ± 0.457 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
