
Beihai sobemo-like virus 22
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEB1|A0A1L3KEB1_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 22 OX=1922694 PE=4 SV=1
MM1 pKa = 8.05DD2 pKa = 4.36WNSSPGVEE10 pKa = 4.0LEE12 pKa = 4.82KK13 pKa = 11.23YY14 pKa = 10.54GSTNRR19 pKa = 11.84QVWQVEE25 pKa = 4.1NGVPDD30 pKa = 4.21PQQKK34 pKa = 10.78LLMWKK39 pKa = 10.03IIQEE43 pKa = 4.35RR44 pKa = 11.84IKK46 pKa = 10.24EE47 pKa = 4.07LQIEE51 pKa = 4.36PTLGPVKK58 pKa = 10.42LFPKK62 pKa = 10.21YY63 pKa = 9.74EE64 pKa = 3.71PHH66 pKa = 6.91KK67 pKa = 9.89ISKK70 pKa = 9.56IRR72 pKa = 11.84EE73 pKa = 3.67NRR75 pKa = 11.84EE76 pKa = 3.39RR77 pKa = 11.84LIFGVGAVDD86 pKa = 4.15NIIAAMFFRR95 pKa = 11.84NWTAALVEE103 pKa = 4.34KK104 pKa = 9.98WRR106 pKa = 11.84EE107 pKa = 3.54IPIKK111 pKa = 10.5VGYY114 pKa = 9.83SPSQGGYY121 pKa = 9.05YY122 pKa = 10.51DD123 pKa = 3.88LFSYY127 pKa = 10.53VSRR130 pKa = 11.84LTNKK134 pKa = 8.2FTAADD139 pKa = 3.54KK140 pKa = 11.02SQWDD144 pKa = 3.69WSCQEE149 pKa = 3.79WEE151 pKa = 4.01VDD153 pKa = 3.44VARR156 pKa = 11.84QVLQHH161 pKa = 6.78LGDD164 pKa = 4.36FEE166 pKa = 7.1DD167 pKa = 4.77GTIADD172 pKa = 4.23NIIRR176 pKa = 11.84AHH178 pKa = 6.86FGPATLVLGKK188 pKa = 10.63AEE190 pKa = 4.18IEE192 pKa = 4.17KK193 pKa = 10.46KK194 pKa = 10.58GYY196 pKa = 10.71GVMFSGTYY204 pKa = 7.79FTLTFNSLFQLILHH218 pKa = 6.73EE219 pKa = 4.59AACVMIGRR227 pKa = 11.84EE228 pKa = 4.0YY229 pKa = 10.84AAPACIGDD237 pKa = 3.95DD238 pKa = 3.63TVQPRR243 pKa = 11.84CEE245 pKa = 4.86DD246 pKa = 3.35EE247 pKa = 4.55QYY249 pKa = 10.51WNAIRR254 pKa = 11.84GFGHH258 pKa = 6.5KK259 pKa = 10.34LKK261 pKa = 10.71DD262 pKa = 3.36IVHH265 pKa = 5.6TTDD268 pKa = 2.73EE269 pKa = 4.75FEE271 pKa = 4.4FAGHH275 pKa = 6.58RR276 pKa = 11.84VTNKK280 pKa = 10.01GAWPVYY286 pKa = 9.5LGKK289 pKa = 9.92HH290 pKa = 4.97RR291 pKa = 11.84AQLQFLTDD299 pKa = 3.52EE300 pKa = 4.49PEE302 pKa = 3.74IRR304 pKa = 11.84MMTLASYY311 pKa = 10.2QRR313 pKa = 11.84LYY315 pKa = 11.28ACDD318 pKa = 3.81ADD320 pKa = 4.2RR321 pKa = 11.84LKK323 pKa = 10.29TFTMALRR330 pKa = 11.84GSQYY334 pKa = 10.34YY335 pKa = 10.5SSSQEE340 pKa = 3.27IRR342 pKa = 11.84NWYY345 pKa = 9.16HH346 pKa = 4.95GWEE349 pKa = 4.13SAANQEE355 pKa = 4.62TAWW358 pKa = 3.65
MM1 pKa = 8.05DD2 pKa = 4.36WNSSPGVEE10 pKa = 4.0LEE12 pKa = 4.82KK13 pKa = 11.23YY14 pKa = 10.54GSTNRR19 pKa = 11.84QVWQVEE25 pKa = 4.1NGVPDD30 pKa = 4.21PQQKK34 pKa = 10.78LLMWKK39 pKa = 10.03IIQEE43 pKa = 4.35RR44 pKa = 11.84IKK46 pKa = 10.24EE47 pKa = 4.07LQIEE51 pKa = 4.36PTLGPVKK58 pKa = 10.42LFPKK62 pKa = 10.21YY63 pKa = 9.74EE64 pKa = 3.71PHH66 pKa = 6.91KK67 pKa = 9.89ISKK70 pKa = 9.56IRR72 pKa = 11.84EE73 pKa = 3.67NRR75 pKa = 11.84EE76 pKa = 3.39RR77 pKa = 11.84LIFGVGAVDD86 pKa = 4.15NIIAAMFFRR95 pKa = 11.84NWTAALVEE103 pKa = 4.34KK104 pKa = 9.98WRR106 pKa = 11.84EE107 pKa = 3.54IPIKK111 pKa = 10.5VGYY114 pKa = 9.83SPSQGGYY121 pKa = 9.05YY122 pKa = 10.51DD123 pKa = 3.88LFSYY127 pKa = 10.53VSRR130 pKa = 11.84LTNKK134 pKa = 8.2FTAADD139 pKa = 3.54KK140 pKa = 11.02SQWDD144 pKa = 3.69WSCQEE149 pKa = 3.79WEE151 pKa = 4.01VDD153 pKa = 3.44VARR156 pKa = 11.84QVLQHH161 pKa = 6.78LGDD164 pKa = 4.36FEE166 pKa = 7.1DD167 pKa = 4.77GTIADD172 pKa = 4.23NIIRR176 pKa = 11.84AHH178 pKa = 6.86FGPATLVLGKK188 pKa = 10.63AEE190 pKa = 4.18IEE192 pKa = 4.17KK193 pKa = 10.46KK194 pKa = 10.58GYY196 pKa = 10.71GVMFSGTYY204 pKa = 7.79FTLTFNSLFQLILHH218 pKa = 6.73EE219 pKa = 4.59AACVMIGRR227 pKa = 11.84EE228 pKa = 4.0YY229 pKa = 10.84AAPACIGDD237 pKa = 3.95DD238 pKa = 3.63TVQPRR243 pKa = 11.84CEE245 pKa = 4.86DD246 pKa = 3.35EE247 pKa = 4.55QYY249 pKa = 10.51WNAIRR254 pKa = 11.84GFGHH258 pKa = 6.5KK259 pKa = 10.34LKK261 pKa = 10.71DD262 pKa = 3.36IVHH265 pKa = 5.6TTDD268 pKa = 2.73EE269 pKa = 4.75FEE271 pKa = 4.4FAGHH275 pKa = 6.58RR276 pKa = 11.84VTNKK280 pKa = 10.01GAWPVYY286 pKa = 9.5LGKK289 pKa = 9.92HH290 pKa = 4.97RR291 pKa = 11.84AQLQFLTDD299 pKa = 3.52EE300 pKa = 4.49PEE302 pKa = 3.74IRR304 pKa = 11.84MMTLASYY311 pKa = 10.2QRR313 pKa = 11.84LYY315 pKa = 11.28ACDD318 pKa = 3.81ADD320 pKa = 4.2RR321 pKa = 11.84LKK323 pKa = 10.29TFTMALRR330 pKa = 11.84GSQYY334 pKa = 10.34YY335 pKa = 10.5SSSQEE340 pKa = 3.27IRR342 pKa = 11.84NWYY345 pKa = 9.16HH346 pKa = 4.95GWEE349 pKa = 4.13SAANQEE355 pKa = 4.62TAWW358 pKa = 3.65
Molecular weight: 41.32 kDa
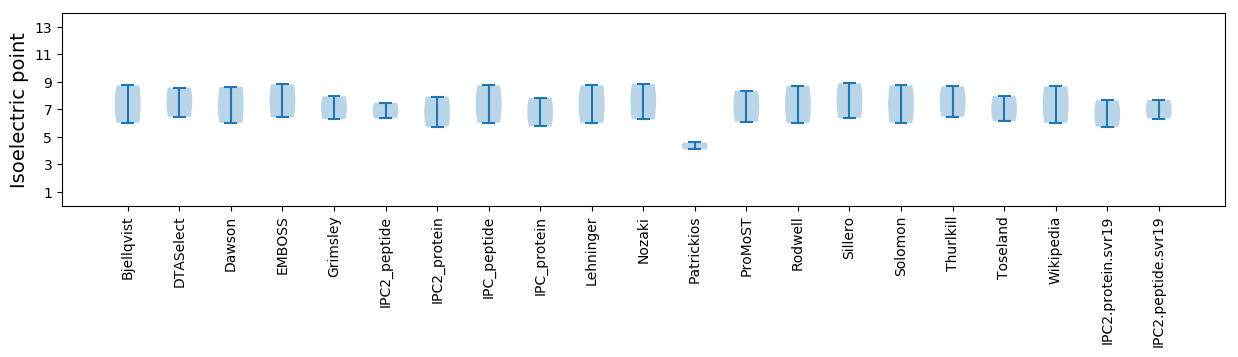
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEB1|A0A1L3KEB1_9VIRU Uncharacterized protein OS=Beihai sobemo-like virus 22 OX=1922694 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 10.52NVLLVLLVVRR12 pKa = 11.84VVVCPNLTLDD22 pKa = 4.21DD23 pKa = 4.53LRR25 pKa = 11.84PIPEE29 pKa = 4.06QLMNMTVDD37 pKa = 2.77AWKK40 pKa = 10.85VYY42 pKa = 7.49MAYY45 pKa = 10.27YY46 pKa = 10.22RR47 pKa = 11.84SPAFNVVRR55 pKa = 11.84CYY57 pKa = 10.97GPWVTFMITVGACAIALLKK76 pKa = 10.42EE77 pKa = 4.14RR78 pKa = 11.84EE79 pKa = 4.11ARR81 pKa = 11.84EE82 pKa = 3.95AKK84 pKa = 10.44AKK86 pKa = 10.56LEE88 pKa = 3.97RR89 pKa = 11.84LFKK92 pKa = 10.58TMHH95 pKa = 5.9TVNGSLRR102 pKa = 11.84CPVLTPEE109 pKa = 4.56SLRR112 pKa = 11.84QGSEE116 pKa = 2.87ISRR119 pKa = 11.84NVKK122 pKa = 9.81KK123 pKa = 11.18VPGTFAVAMKK133 pKa = 10.41LAGEE137 pKa = 4.24LTVVGYY143 pKa = 9.67GMRR146 pKa = 11.84LEE148 pKa = 4.29NAAVFPLHH156 pKa = 6.69LCGDD160 pKa = 4.25SEE162 pKa = 5.47DD163 pKa = 3.9GTITIMNNGKK173 pKa = 9.77NGLKK177 pKa = 9.25TATLEE182 pKa = 4.08YY183 pKa = 9.49SAHH186 pKa = 6.24VEE188 pKa = 3.6VDD190 pKa = 2.86TDD192 pKa = 3.55VGVWQKK198 pKa = 7.66TTDD201 pKa = 3.49FWSQLEE207 pKa = 4.21VGKK210 pKa = 10.89ANIGILSAPAMAQAIAGGKK229 pKa = 8.91MSMGTLKK236 pKa = 10.57NDD238 pKa = 3.55PEE240 pKa = 4.5VVGRR244 pKa = 11.84LEE246 pKa = 4.14YY247 pKa = 10.78AGSTEE252 pKa = 4.22AGFSGGAYY260 pKa = 10.01LVATQIVGMHH270 pKa = 6.21FSGGSVNKK278 pKa = 10.12GFALMYY284 pKa = 9.6LYY286 pKa = 11.11SLIRR290 pKa = 11.84SVFNQEE296 pKa = 3.86AGSDD300 pKa = 4.06YY301 pKa = 11.31SWMNALVKK309 pKa = 10.56RR310 pKa = 11.84KK311 pKa = 9.72VKK313 pKa = 10.1VRR315 pKa = 11.84ARR317 pKa = 11.84RR318 pKa = 11.84HH319 pKa = 4.24PTNRR323 pKa = 11.84DD324 pKa = 3.16EE325 pKa = 6.62VIMQYY330 pKa = 10.32EE331 pKa = 4.56GQYY334 pKa = 10.86HH335 pKa = 6.09LVPYY339 pKa = 10.12EE340 pKa = 4.17EE341 pKa = 4.33FSMRR345 pKa = 11.84YY346 pKa = 8.12GALGAGEE353 pKa = 4.14TDD355 pKa = 3.61FVGRR359 pKa = 11.84EE360 pKa = 4.07VVPEE364 pKa = 4.33VYY366 pKa = 9.66TGHH369 pKa = 6.96TDD371 pKa = 3.95NIAGDD376 pKa = 3.92LAFNTAVPLNYY387 pKa = 10.04DD388 pKa = 3.72NPQVTPGGPHH398 pKa = 5.11RR399 pKa = 11.84TKK401 pKa = 10.61NAEE404 pKa = 4.13NVEE407 pKa = 4.17KK408 pKa = 9.48TLQMNALEE416 pKa = 4.47LQLSKK421 pKa = 10.92LRR423 pKa = 11.84EE424 pKa = 3.89KK425 pKa = 11.3YY426 pKa = 9.55EE427 pKa = 4.53AIVRR431 pKa = 11.84ATEE434 pKa = 3.99KK435 pKa = 10.5FVAQYY440 pKa = 10.89QPDD443 pKa = 3.95PKK445 pKa = 10.89AKK447 pKa = 10.63EE448 pKa = 3.54EE449 pKa = 4.01DD450 pKa = 3.19ALLRR454 pKa = 11.84GKK456 pKa = 9.18MKK458 pKa = 10.36QAAQEE463 pKa = 3.89ITAEE467 pKa = 3.83IHH469 pKa = 6.63KK470 pKa = 10.61EE471 pKa = 3.63MEE473 pKa = 4.38AVSKK477 pKa = 10.97SLTEE481 pKa = 3.82VRR483 pKa = 11.84RR484 pKa = 11.84NTDD487 pKa = 2.88EE488 pKa = 4.3EE489 pKa = 3.83IRR491 pKa = 11.84NRR493 pKa = 11.84RR494 pKa = 11.84RR495 pKa = 11.84AKK497 pKa = 9.04KK498 pKa = 7.9TVRR501 pKa = 11.84KK502 pKa = 8.86LASNEE507 pKa = 3.74DD508 pKa = 3.56VVRR511 pKa = 11.84NSLQRR516 pKa = 11.84VPDD519 pKa = 4.42FVQCHH524 pKa = 5.88FGDD527 pKa = 4.06LPTEE531 pKa = 3.88LRR533 pKa = 11.84DD534 pKa = 3.65RR535 pKa = 11.84LKK537 pKa = 11.21DD538 pKa = 3.41STTSILKK545 pKa = 10.43LKK547 pKa = 10.78VPTTSTSSQRR557 pKa = 11.84SPEE560 pKa = 3.85NDD562 pKa = 3.25TPLPSAGTPLSPPSS576 pKa = 3.39
MM1 pKa = 7.45KK2 pKa = 10.52NVLLVLLVVRR12 pKa = 11.84VVVCPNLTLDD22 pKa = 4.21DD23 pKa = 4.53LRR25 pKa = 11.84PIPEE29 pKa = 4.06QLMNMTVDD37 pKa = 2.77AWKK40 pKa = 10.85VYY42 pKa = 7.49MAYY45 pKa = 10.27YY46 pKa = 10.22RR47 pKa = 11.84SPAFNVVRR55 pKa = 11.84CYY57 pKa = 10.97GPWVTFMITVGACAIALLKK76 pKa = 10.42EE77 pKa = 4.14RR78 pKa = 11.84EE79 pKa = 4.11ARR81 pKa = 11.84EE82 pKa = 3.95AKK84 pKa = 10.44AKK86 pKa = 10.56LEE88 pKa = 3.97RR89 pKa = 11.84LFKK92 pKa = 10.58TMHH95 pKa = 5.9TVNGSLRR102 pKa = 11.84CPVLTPEE109 pKa = 4.56SLRR112 pKa = 11.84QGSEE116 pKa = 2.87ISRR119 pKa = 11.84NVKK122 pKa = 9.81KK123 pKa = 11.18VPGTFAVAMKK133 pKa = 10.41LAGEE137 pKa = 4.24LTVVGYY143 pKa = 9.67GMRR146 pKa = 11.84LEE148 pKa = 4.29NAAVFPLHH156 pKa = 6.69LCGDD160 pKa = 4.25SEE162 pKa = 5.47DD163 pKa = 3.9GTITIMNNGKK173 pKa = 9.77NGLKK177 pKa = 9.25TATLEE182 pKa = 4.08YY183 pKa = 9.49SAHH186 pKa = 6.24VEE188 pKa = 3.6VDD190 pKa = 2.86TDD192 pKa = 3.55VGVWQKK198 pKa = 7.66TTDD201 pKa = 3.49FWSQLEE207 pKa = 4.21VGKK210 pKa = 10.89ANIGILSAPAMAQAIAGGKK229 pKa = 8.91MSMGTLKK236 pKa = 10.57NDD238 pKa = 3.55PEE240 pKa = 4.5VVGRR244 pKa = 11.84LEE246 pKa = 4.14YY247 pKa = 10.78AGSTEE252 pKa = 4.22AGFSGGAYY260 pKa = 10.01LVATQIVGMHH270 pKa = 6.21FSGGSVNKK278 pKa = 10.12GFALMYY284 pKa = 9.6LYY286 pKa = 11.11SLIRR290 pKa = 11.84SVFNQEE296 pKa = 3.86AGSDD300 pKa = 4.06YY301 pKa = 11.31SWMNALVKK309 pKa = 10.56RR310 pKa = 11.84KK311 pKa = 9.72VKK313 pKa = 10.1VRR315 pKa = 11.84ARR317 pKa = 11.84RR318 pKa = 11.84HH319 pKa = 4.24PTNRR323 pKa = 11.84DD324 pKa = 3.16EE325 pKa = 6.62VIMQYY330 pKa = 10.32EE331 pKa = 4.56GQYY334 pKa = 10.86HH335 pKa = 6.09LVPYY339 pKa = 10.12EE340 pKa = 4.17EE341 pKa = 4.33FSMRR345 pKa = 11.84YY346 pKa = 8.12GALGAGEE353 pKa = 4.14TDD355 pKa = 3.61FVGRR359 pKa = 11.84EE360 pKa = 4.07VVPEE364 pKa = 4.33VYY366 pKa = 9.66TGHH369 pKa = 6.96TDD371 pKa = 3.95NIAGDD376 pKa = 3.92LAFNTAVPLNYY387 pKa = 10.04DD388 pKa = 3.72NPQVTPGGPHH398 pKa = 5.11RR399 pKa = 11.84TKK401 pKa = 10.61NAEE404 pKa = 4.13NVEE407 pKa = 4.17KK408 pKa = 9.48TLQMNALEE416 pKa = 4.47LQLSKK421 pKa = 10.92LRR423 pKa = 11.84EE424 pKa = 3.89KK425 pKa = 11.3YY426 pKa = 9.55EE427 pKa = 4.53AIVRR431 pKa = 11.84ATEE434 pKa = 3.99KK435 pKa = 10.5FVAQYY440 pKa = 10.89QPDD443 pKa = 3.95PKK445 pKa = 10.89AKK447 pKa = 10.63EE448 pKa = 3.54EE449 pKa = 4.01DD450 pKa = 3.19ALLRR454 pKa = 11.84GKK456 pKa = 9.18MKK458 pKa = 10.36QAAQEE463 pKa = 3.89ITAEE467 pKa = 3.83IHH469 pKa = 6.63KK470 pKa = 10.61EE471 pKa = 3.63MEE473 pKa = 4.38AVSKK477 pKa = 10.97SLTEE481 pKa = 3.82VRR483 pKa = 11.84RR484 pKa = 11.84NTDD487 pKa = 2.88EE488 pKa = 4.3EE489 pKa = 3.83IRR491 pKa = 11.84NRR493 pKa = 11.84RR494 pKa = 11.84RR495 pKa = 11.84AKK497 pKa = 9.04KK498 pKa = 7.9TVRR501 pKa = 11.84KK502 pKa = 8.86LASNEE507 pKa = 3.74DD508 pKa = 3.56VVRR511 pKa = 11.84NSLQRR516 pKa = 11.84VPDD519 pKa = 4.42FVQCHH524 pKa = 5.88FGDD527 pKa = 4.06LPTEE531 pKa = 3.88LRR533 pKa = 11.84DD534 pKa = 3.65RR535 pKa = 11.84LKK537 pKa = 11.21DD538 pKa = 3.41STTSILKK545 pKa = 10.43LKK547 pKa = 10.78VPTTSTSSQRR557 pKa = 11.84SPEE560 pKa = 3.85NDD562 pKa = 3.25TPLPSAGTPLSPPSS576 pKa = 3.39
Molecular weight: 63.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
934 |
358 |
576 |
467.0 |
52.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.244 ± 0.234 | 1.178 ± 0.121 |
4.497 ± 0.295 | 7.388 ± 0.086 |
3.533 ± 0.674 | 6.959 ± 0.013 |
2.034 ± 0.266 | 4.283 ± 1.033 |
5.996 ± 0.227 | 8.351 ± 0.604 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.998 ± 0.423 | 4.39 ± 0.421 |
4.604 ± 0.384 | 4.176 ± 0.782 |
5.996 ± 0.227 | 5.46 ± 0.24 |
6.317 ± 0.56 | 7.923 ± 1.296 |
1.927 ± 0.945 | 3.747 ± 0.4 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
