
Equus caballus papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyoiotapapillomavirus; Dyoiotapapillomavirus 1
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
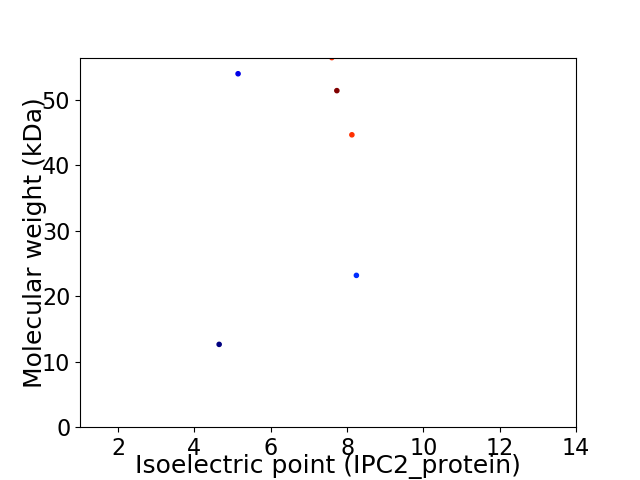
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
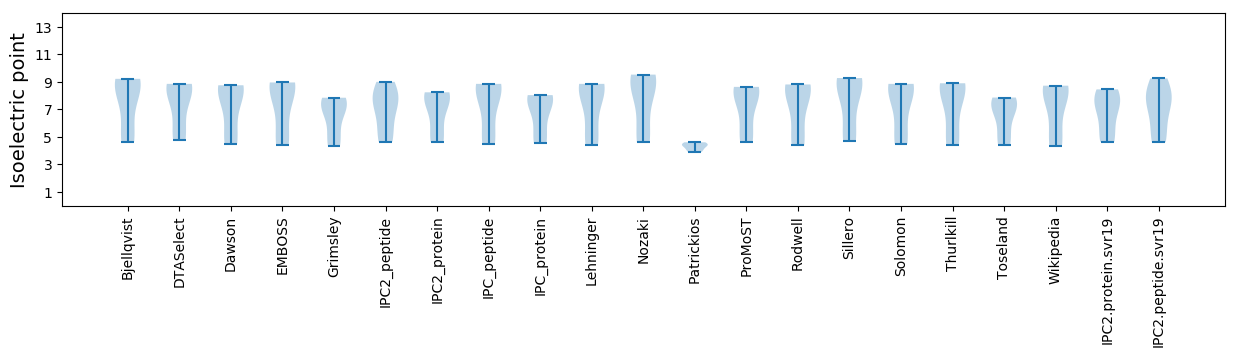
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C0ITA7|C0ITA7_9PAPI Putative E1 OS=Equus caballus papillomavirus 2 OX=526413 PE=4 SV=1
MM1 pKa = 7.75RR2 pKa = 11.84GLHH5 pKa = 6.61RR6 pKa = 11.84LLPDD10 pKa = 3.28VVLDD14 pKa = 3.92ALSSITLAEE23 pKa = 4.16RR24 pKa = 11.84SPSVSSSDD32 pKa = 3.66TEE34 pKa = 4.6DD35 pKa = 5.42EE36 pKa = 3.94EE37 pKa = 4.45DD38 pKa = 3.91QPDD41 pKa = 3.62GGAQRR46 pKa = 11.84VAEE49 pKa = 4.27EE50 pKa = 4.11DD51 pKa = 3.86LEE53 pKa = 4.79VEE55 pKa = 4.62VVPRR59 pKa = 11.84EE60 pKa = 4.07HH61 pKa = 6.19PQFRR65 pKa = 11.84VLTLCEE71 pKa = 3.68RR72 pKa = 11.84CNRR75 pKa = 11.84GVRR78 pKa = 11.84LVVVGPTGDD87 pKa = 3.54VRR89 pKa = 11.84ILQQLLLGSLGIVCADD105 pKa = 3.76CASQEE110 pKa = 3.93GHH112 pKa = 6.38NGHH115 pKa = 6.12GQQ117 pKa = 3.03
MM1 pKa = 7.75RR2 pKa = 11.84GLHH5 pKa = 6.61RR6 pKa = 11.84LLPDD10 pKa = 3.28VVLDD14 pKa = 3.92ALSSITLAEE23 pKa = 4.16RR24 pKa = 11.84SPSVSSSDD32 pKa = 3.66TEE34 pKa = 4.6DD35 pKa = 5.42EE36 pKa = 3.94EE37 pKa = 4.45DD38 pKa = 3.91QPDD41 pKa = 3.62GGAQRR46 pKa = 11.84VAEE49 pKa = 4.27EE50 pKa = 4.11DD51 pKa = 3.86LEE53 pKa = 4.79VEE55 pKa = 4.62VVPRR59 pKa = 11.84EE60 pKa = 4.07HH61 pKa = 6.19PQFRR65 pKa = 11.84VLTLCEE71 pKa = 3.68RR72 pKa = 11.84CNRR75 pKa = 11.84GVRR78 pKa = 11.84LVVVGPTGDD87 pKa = 3.54VRR89 pKa = 11.84ILQQLLLGSLGIVCADD105 pKa = 3.76CASQEE110 pKa = 3.93GHH112 pKa = 6.38NGHH115 pKa = 6.12GQQ117 pKa = 3.03
Molecular weight: 12.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0ITA6|C0ITA6_9PAPI Protein E7 OS=Equus caballus papillomavirus 2 OX=526413 GN=E7 PE=3 SV=1
MM1 pKa = 7.98PSRR4 pKa = 11.84PVPAKK9 pKa = 10.79SFGALFWLLWRR20 pKa = 11.84CTQPICRR27 pKa = 11.84LGLCEE32 pKa = 3.89SANPLRR38 pKa = 11.84CTEE41 pKa = 3.89PFSRR45 pKa = 11.84EE46 pKa = 4.17DD47 pKa = 3.27CCGPQSLRR55 pKa = 11.84YY56 pKa = 9.88AGGPCIGDD64 pKa = 3.31VPRR67 pKa = 11.84SSRR70 pKa = 11.84GRR72 pKa = 11.84VKK74 pKa = 9.99TAGHH78 pKa = 5.6MSIVGLLCIFCGKK91 pKa = 8.88PCGYY95 pKa = 10.94ADD97 pKa = 5.19DD98 pKa = 4.54IWWNTADD105 pKa = 4.37LNLVMRR111 pKa = 11.84DD112 pKa = 3.6GKK114 pKa = 10.76RR115 pKa = 11.84KK116 pKa = 9.8ASCQTCMRR124 pKa = 11.84AACRR128 pKa = 11.84LEE130 pKa = 3.56RR131 pKa = 11.84HH132 pKa = 6.6LYY134 pKa = 8.19QSSRR138 pKa = 11.84NVYY141 pKa = 6.53TARR144 pKa = 11.84GVEE147 pKa = 4.29RR148 pKa = 11.84ACGQSVYY155 pKa = 10.04RR156 pKa = 11.84VSVRR160 pKa = 11.84CTHH163 pKa = 6.09CGKK166 pKa = 10.53RR167 pKa = 11.84LTRR170 pKa = 11.84DD171 pKa = 3.06EE172 pKa = 4.38KK173 pKa = 10.7RR174 pKa = 11.84WIEE177 pKa = 3.65IEE179 pKa = 3.98GAGFYY184 pKa = 8.33QVRR187 pKa = 11.84QRR189 pKa = 11.84WRR191 pKa = 11.84ALCYY195 pKa = 10.07DD196 pKa = 4.1CRR198 pKa = 11.84VCDD201 pKa = 3.88EE202 pKa = 4.58GSAA205 pKa = 3.53
MM1 pKa = 7.98PSRR4 pKa = 11.84PVPAKK9 pKa = 10.79SFGALFWLLWRR20 pKa = 11.84CTQPICRR27 pKa = 11.84LGLCEE32 pKa = 3.89SANPLRR38 pKa = 11.84CTEE41 pKa = 3.89PFSRR45 pKa = 11.84EE46 pKa = 4.17DD47 pKa = 3.27CCGPQSLRR55 pKa = 11.84YY56 pKa = 9.88AGGPCIGDD64 pKa = 3.31VPRR67 pKa = 11.84SSRR70 pKa = 11.84GRR72 pKa = 11.84VKK74 pKa = 9.99TAGHH78 pKa = 5.6MSIVGLLCIFCGKK91 pKa = 8.88PCGYY95 pKa = 10.94ADD97 pKa = 5.19DD98 pKa = 4.54IWWNTADD105 pKa = 4.37LNLVMRR111 pKa = 11.84DD112 pKa = 3.6GKK114 pKa = 10.76RR115 pKa = 11.84KK116 pKa = 9.8ASCQTCMRR124 pKa = 11.84AACRR128 pKa = 11.84LEE130 pKa = 3.56RR131 pKa = 11.84HH132 pKa = 6.6LYY134 pKa = 8.19QSSRR138 pKa = 11.84NVYY141 pKa = 6.53TARR144 pKa = 11.84GVEE147 pKa = 4.29RR148 pKa = 11.84ACGQSVYY155 pKa = 10.04RR156 pKa = 11.84VSVRR160 pKa = 11.84CTHH163 pKa = 6.09CGKK166 pKa = 10.53RR167 pKa = 11.84LTRR170 pKa = 11.84DD171 pKa = 3.06EE172 pKa = 4.38KK173 pKa = 10.7RR174 pKa = 11.84WIEE177 pKa = 3.65IEE179 pKa = 3.98GAGFYY184 pKa = 8.33QVRR187 pKa = 11.84QRR189 pKa = 11.84WRR191 pKa = 11.84ALCYY195 pKa = 10.07DD196 pKa = 4.1CRR198 pKa = 11.84VCDD201 pKa = 3.88EE202 pKa = 4.58GSAA205 pKa = 3.53
Molecular weight: 23.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2200 |
117 |
519 |
366.7 |
40.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.045 ± 0.553 | 2.5 ± 0.909 |
5.591 ± 0.282 | 5.227 ± 0.406 |
4.591 ± 0.54 | 9.318 ± 1.718 |
2.5 ± 0.367 | 2.409 ± 0.409 |
4.091 ± 0.909 | 8.455 ± 0.719 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.227 ± 0.548 | 3.227 ± 0.745 |
6.682 ± 0.604 | 4.182 ± 0.215 |
7.227 ± 0.68 | 7.636 ± 0.778 |
5.182 ± 0.262 | 7.955 ± 0.781 |
1.455 ± 0.454 | 2.5 ± 0.438 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
