
Ctenophore-associated circular genome 4
Taxonomy: Viruses; Satellites; DNA satellites; Single stranded DNA satellites
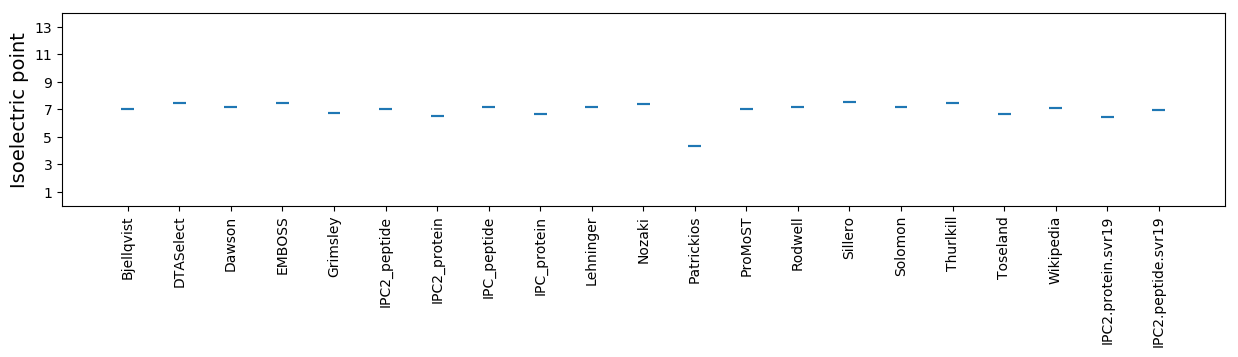
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
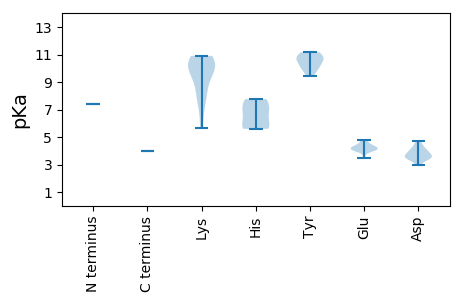
>tr|A0A141MJB6|A0A141MJB6_9VIRU ATP-dependent helicase Rep OS=Ctenophore-associated circular genome 4 OX=1778573 PE=3 SV=1
MM1 pKa = 7.4VRR3 pKa = 11.84EE4 pKa = 4.09SAAVGWCFTLNNPTDD19 pKa = 3.79EE20 pKa = 4.61EE21 pKa = 4.42VDD23 pKa = 3.44NLNRR27 pKa = 11.84ILDD30 pKa = 4.04GDD32 pKa = 3.9QIKK35 pKa = 10.32YY36 pKa = 10.59AIYY39 pKa = 10.06QKK41 pKa = 9.61EE42 pKa = 4.18TGTPHH47 pKa = 5.6LQGYY51 pKa = 9.44LEE53 pKa = 4.22VSRR56 pKa = 11.84KK57 pKa = 7.75QRR59 pKa = 11.84LVGVKK64 pKa = 10.1RR65 pKa = 11.84LLQTEE70 pKa = 4.27RR71 pKa = 11.84VHH73 pKa = 7.75LEE75 pKa = 3.5KK76 pKa = 10.81RR77 pKa = 11.84KK78 pKa = 8.48GTRR81 pKa = 11.84IQARR85 pKa = 11.84DD86 pKa = 3.54YY87 pKa = 10.96CRR89 pKa = 11.84KK90 pKa = 8.4TEE92 pKa = 4.01GRR94 pKa = 11.84LADD97 pKa = 3.54PVVFGVWRR105 pKa = 11.84GDD107 pKa = 3.54SQTTGMSRR115 pKa = 11.84VIARR119 pKa = 11.84LEE121 pKa = 4.39GGCTLSEE128 pKa = 4.36VAAEE132 pKa = 4.19EE133 pKa = 4.22PQTFIQYY140 pKa = 10.77HH141 pKa = 5.66NGLAKK146 pKa = 9.92WYY148 pKa = 10.59ALVCTFARR156 pKa = 11.84EE157 pKa = 3.94TDD159 pKa = 3.42SPRR162 pKa = 11.84GVWIYY167 pKa = 10.6GPPGIGKK174 pKa = 5.65THH176 pKa = 7.62KK177 pKa = 10.51ARR179 pKa = 11.84ALASEE184 pKa = 4.07SLYY187 pKa = 11.13LKK189 pKa = 10.32SQNKK193 pKa = 7.34WWDD196 pKa = 3.47GYY198 pKa = 11.17AGQKK202 pKa = 10.02FVVLDD207 pKa = 3.81DD208 pKa = 4.44FDD210 pKa = 4.23RR211 pKa = 11.84QGICLGHH218 pKa = 5.82YY219 pKa = 10.12LKK221 pKa = 10.23IWSDD225 pKa = 2.95KK226 pKa = 9.4WSCTAEE232 pKa = 4.01IKK234 pKa = 10.9GGTVNLQHH242 pKa = 6.92EE243 pKa = 4.76KK244 pKa = 10.82FIVTSNYY251 pKa = 9.94HH252 pKa = 6.82PDD254 pKa = 3.88EE255 pKa = 4.77LWTDD259 pKa = 3.62DD260 pKa = 4.19PQLVDD265 pKa = 4.73AISRR269 pKa = 11.84RR270 pKa = 11.84FEE272 pKa = 4.34IIRR275 pKa = 11.84GEE277 pKa = 4.01SDD279 pKa = 3.41TNVTNWIVEE288 pKa = 4.11NN289 pKa = 4.0
MM1 pKa = 7.4VRR3 pKa = 11.84EE4 pKa = 4.09SAAVGWCFTLNNPTDD19 pKa = 3.79EE20 pKa = 4.61EE21 pKa = 4.42VDD23 pKa = 3.44NLNRR27 pKa = 11.84ILDD30 pKa = 4.04GDD32 pKa = 3.9QIKK35 pKa = 10.32YY36 pKa = 10.59AIYY39 pKa = 10.06QKK41 pKa = 9.61EE42 pKa = 4.18TGTPHH47 pKa = 5.6LQGYY51 pKa = 9.44LEE53 pKa = 4.22VSRR56 pKa = 11.84KK57 pKa = 7.75QRR59 pKa = 11.84LVGVKK64 pKa = 10.1RR65 pKa = 11.84LLQTEE70 pKa = 4.27RR71 pKa = 11.84VHH73 pKa = 7.75LEE75 pKa = 3.5KK76 pKa = 10.81RR77 pKa = 11.84KK78 pKa = 8.48GTRR81 pKa = 11.84IQARR85 pKa = 11.84DD86 pKa = 3.54YY87 pKa = 10.96CRR89 pKa = 11.84KK90 pKa = 8.4TEE92 pKa = 4.01GRR94 pKa = 11.84LADD97 pKa = 3.54PVVFGVWRR105 pKa = 11.84GDD107 pKa = 3.54SQTTGMSRR115 pKa = 11.84VIARR119 pKa = 11.84LEE121 pKa = 4.39GGCTLSEE128 pKa = 4.36VAAEE132 pKa = 4.19EE133 pKa = 4.22PQTFIQYY140 pKa = 10.77HH141 pKa = 5.66NGLAKK146 pKa = 9.92WYY148 pKa = 10.59ALVCTFARR156 pKa = 11.84EE157 pKa = 3.94TDD159 pKa = 3.42SPRR162 pKa = 11.84GVWIYY167 pKa = 10.6GPPGIGKK174 pKa = 5.65THH176 pKa = 7.62KK177 pKa = 10.51ARR179 pKa = 11.84ALASEE184 pKa = 4.07SLYY187 pKa = 11.13LKK189 pKa = 10.32SQNKK193 pKa = 7.34WWDD196 pKa = 3.47GYY198 pKa = 11.17AGQKK202 pKa = 10.02FVVLDD207 pKa = 3.81DD208 pKa = 4.44FDD210 pKa = 4.23RR211 pKa = 11.84QGICLGHH218 pKa = 5.82YY219 pKa = 10.12LKK221 pKa = 10.23IWSDD225 pKa = 2.95KK226 pKa = 9.4WSCTAEE232 pKa = 4.01IKK234 pKa = 10.9GGTVNLQHH242 pKa = 6.92EE243 pKa = 4.76KK244 pKa = 10.82FIVTSNYY251 pKa = 9.94HH252 pKa = 6.82PDD254 pKa = 3.88EE255 pKa = 4.77LWTDD259 pKa = 3.62DD260 pKa = 4.19PQLVDD265 pKa = 4.73AISRR269 pKa = 11.84RR270 pKa = 11.84FEE272 pKa = 4.34IIRR275 pKa = 11.84GEE277 pKa = 4.01SDD279 pKa = 3.41TNVTNWIVEE288 pKa = 4.11NN289 pKa = 4.0
Molecular weight: 33.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A141MJB6|A0A141MJB6_9VIRU ATP-dependent helicase Rep OS=Ctenophore-associated circular genome 4 OX=1778573 PE=3 SV=1
MM1 pKa = 7.4VRR3 pKa = 11.84EE4 pKa = 4.09SAAVGWCFTLNNPTDD19 pKa = 3.79EE20 pKa = 4.61EE21 pKa = 4.42VDD23 pKa = 3.44NLNRR27 pKa = 11.84ILDD30 pKa = 4.04GDD32 pKa = 3.9QIKK35 pKa = 10.32YY36 pKa = 10.59AIYY39 pKa = 10.06QKK41 pKa = 9.61EE42 pKa = 4.18TGTPHH47 pKa = 5.6LQGYY51 pKa = 9.44LEE53 pKa = 4.22VSRR56 pKa = 11.84KK57 pKa = 7.75QRR59 pKa = 11.84LVGVKK64 pKa = 10.1RR65 pKa = 11.84LLQTEE70 pKa = 4.27RR71 pKa = 11.84VHH73 pKa = 7.75LEE75 pKa = 3.5KK76 pKa = 10.81RR77 pKa = 11.84KK78 pKa = 8.48GTRR81 pKa = 11.84IQARR85 pKa = 11.84DD86 pKa = 3.54YY87 pKa = 10.96CRR89 pKa = 11.84KK90 pKa = 8.4TEE92 pKa = 4.01GRR94 pKa = 11.84LADD97 pKa = 3.54PVVFGVWRR105 pKa = 11.84GDD107 pKa = 3.54SQTTGMSRR115 pKa = 11.84VIARR119 pKa = 11.84LEE121 pKa = 4.39GGCTLSEE128 pKa = 4.36VAAEE132 pKa = 4.19EE133 pKa = 4.22PQTFIQYY140 pKa = 10.77HH141 pKa = 5.66NGLAKK146 pKa = 9.92WYY148 pKa = 10.59ALVCTFARR156 pKa = 11.84EE157 pKa = 3.94TDD159 pKa = 3.42SPRR162 pKa = 11.84GVWIYY167 pKa = 10.6GPPGIGKK174 pKa = 5.65THH176 pKa = 7.62KK177 pKa = 10.51ARR179 pKa = 11.84ALASEE184 pKa = 4.07SLYY187 pKa = 11.13LKK189 pKa = 10.32SQNKK193 pKa = 7.34WWDD196 pKa = 3.47GYY198 pKa = 11.17AGQKK202 pKa = 10.02FVVLDD207 pKa = 3.81DD208 pKa = 4.44FDD210 pKa = 4.23RR211 pKa = 11.84QGICLGHH218 pKa = 5.82YY219 pKa = 10.12LKK221 pKa = 10.23IWSDD225 pKa = 2.95KK226 pKa = 9.4WSCTAEE232 pKa = 4.01IKK234 pKa = 10.9GGTVNLQHH242 pKa = 6.92EE243 pKa = 4.76KK244 pKa = 10.82FIVTSNYY251 pKa = 9.94HH252 pKa = 6.82PDD254 pKa = 3.88EE255 pKa = 4.77LWTDD259 pKa = 3.62DD260 pKa = 4.19PQLVDD265 pKa = 4.73AISRR269 pKa = 11.84RR270 pKa = 11.84FEE272 pKa = 4.34IIRR275 pKa = 11.84GEE277 pKa = 4.01SDD279 pKa = 3.41TNVTNWIVEE288 pKa = 4.11NN289 pKa = 4.0
MM1 pKa = 7.4VRR3 pKa = 11.84EE4 pKa = 4.09SAAVGWCFTLNNPTDD19 pKa = 3.79EE20 pKa = 4.61EE21 pKa = 4.42VDD23 pKa = 3.44NLNRR27 pKa = 11.84ILDD30 pKa = 4.04GDD32 pKa = 3.9QIKK35 pKa = 10.32YY36 pKa = 10.59AIYY39 pKa = 10.06QKK41 pKa = 9.61EE42 pKa = 4.18TGTPHH47 pKa = 5.6LQGYY51 pKa = 9.44LEE53 pKa = 4.22VSRR56 pKa = 11.84KK57 pKa = 7.75QRR59 pKa = 11.84LVGVKK64 pKa = 10.1RR65 pKa = 11.84LLQTEE70 pKa = 4.27RR71 pKa = 11.84VHH73 pKa = 7.75LEE75 pKa = 3.5KK76 pKa = 10.81RR77 pKa = 11.84KK78 pKa = 8.48GTRR81 pKa = 11.84IQARR85 pKa = 11.84DD86 pKa = 3.54YY87 pKa = 10.96CRR89 pKa = 11.84KK90 pKa = 8.4TEE92 pKa = 4.01GRR94 pKa = 11.84LADD97 pKa = 3.54PVVFGVWRR105 pKa = 11.84GDD107 pKa = 3.54SQTTGMSRR115 pKa = 11.84VIARR119 pKa = 11.84LEE121 pKa = 4.39GGCTLSEE128 pKa = 4.36VAAEE132 pKa = 4.19EE133 pKa = 4.22PQTFIQYY140 pKa = 10.77HH141 pKa = 5.66NGLAKK146 pKa = 9.92WYY148 pKa = 10.59ALVCTFARR156 pKa = 11.84EE157 pKa = 3.94TDD159 pKa = 3.42SPRR162 pKa = 11.84GVWIYY167 pKa = 10.6GPPGIGKK174 pKa = 5.65THH176 pKa = 7.62KK177 pKa = 10.51ARR179 pKa = 11.84ALASEE184 pKa = 4.07SLYY187 pKa = 11.13LKK189 pKa = 10.32SQNKK193 pKa = 7.34WWDD196 pKa = 3.47GYY198 pKa = 11.17AGQKK202 pKa = 10.02FVVLDD207 pKa = 3.81DD208 pKa = 4.44FDD210 pKa = 4.23RR211 pKa = 11.84QGICLGHH218 pKa = 5.82YY219 pKa = 10.12LKK221 pKa = 10.23IWSDD225 pKa = 2.95KK226 pKa = 9.4WSCTAEE232 pKa = 4.01IKK234 pKa = 10.9GGTVNLQHH242 pKa = 6.92EE243 pKa = 4.76KK244 pKa = 10.82FIVTSNYY251 pKa = 9.94HH252 pKa = 6.82PDD254 pKa = 3.88EE255 pKa = 4.77LWTDD259 pKa = 3.62DD260 pKa = 4.19PQLVDD265 pKa = 4.73AISRR269 pKa = 11.84RR270 pKa = 11.84FEE272 pKa = 4.34IIRR275 pKa = 11.84GEE277 pKa = 4.01SDD279 pKa = 3.41TNVTNWIVEE288 pKa = 4.11NN289 pKa = 4.0
Molecular weight: 33.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
289 |
289 |
289 |
289.0 |
33.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.882 ± 0.0 | 2.076 ± 0.0 |
6.228 ± 0.0 | 6.92 ± 0.0 |
2.768 ± 0.0 | 8.304 ± 0.0 |
2.422 ± 0.0 | 5.536 ± 0.0 |
5.882 ± 0.0 | 7.958 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.692 ± 0.0 | 3.806 ± 0.0 |
3.114 ± 0.0 | 4.844 ± 0.0 |
7.266 ± 0.0 | 4.844 ± 0.0 |
6.92 ± 0.0 | 7.266 ± 0.0 |
3.46 ± 0.0 | 3.806 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
