
Prunus dulcis (Almond) (Amygdalus dulcis)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales;
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
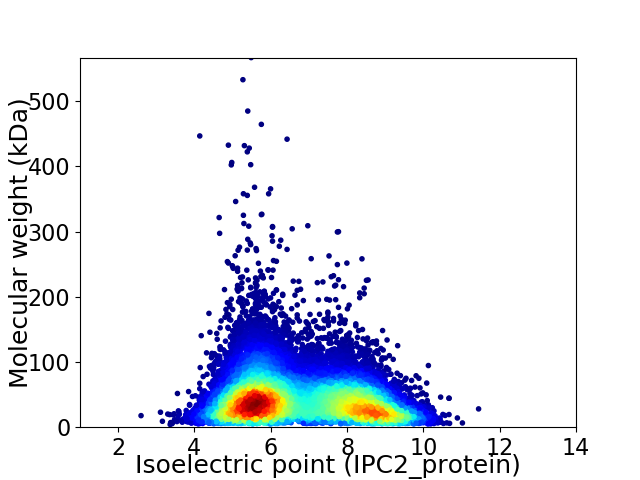
Virtual 2D-PAGE plot for 31934 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5E4E4M8|A0A5E4E4M8_PRUDU Uncharacterized protein OS=Prunus dulcis OX=3755 GN=ALMOND_2B026615 PE=4 SV=1
MM1 pKa = 7.36FPLSKK6 pKa = 10.63LIISASWYY14 pKa = 8.29EE15 pKa = 3.88VHH17 pKa = 7.34GDD19 pKa = 4.04GVDD22 pKa = 3.92DD23 pKa = 5.12FNGDD27 pKa = 4.5DD28 pKa = 3.66IFALAACIEE37 pKa = 4.22GDD39 pKa = 4.59GDD41 pKa = 6.02DD42 pKa = 5.53DD43 pKa = 6.4DD44 pKa = 6.51SDD46 pKa = 4.13YY47 pKa = 11.41DD48 pKa = 3.78YY49 pKa = 11.73APAAMEE55 pKa = 5.19GDD57 pKa = 5.03DD58 pKa = 6.13DD59 pKa = 6.8DD60 pKa = 7.45DD61 pKa = 6.72DD62 pKa = 6.51DD63 pKa = 6.51GEE65 pKa = 4.41IVLPINSVRR74 pKa = 11.84LEE76 pKa = 4.11EE77 pKa = 4.11SSMWKK82 pKa = 10.27DD83 pKa = 3.07KK84 pKa = 11.14YY85 pKa = 11.05SPKK88 pKa = 10.03FDD90 pKa = 3.23GKK92 pKa = 10.18VNRR95 pKa = 11.84VDD97 pKa = 4.37RR98 pKa = 11.84KK99 pKa = 9.6NEE101 pKa = 3.8LYY103 pKa = 10.6ASRR106 pKa = 11.84VHH108 pKa = 5.38QNYY111 pKa = 9.0YY112 pKa = 10.79FMAPVFKK119 pKa = 10.81LIGLSCYY126 pKa = 9.74SNHH129 pKa = 5.34MHH131 pKa = 7.59GDD133 pKa = 3.22GDD135 pKa = 4.93DD136 pKa = 3.57IYY138 pKa = 11.54GPALNGTAMTTTFPTMTTLQAPAACTEE165 pKa = 4.16WEE167 pKa = 4.05GDD169 pKa = 4.15DD170 pKa = 3.9YY171 pKa = 12.09DD172 pKa = 4.13EE173 pKa = 4.59STTILTMILHH183 pKa = 5.29QQHH186 pKa = 7.26DD187 pKa = 4.27LMLIIVVHH195 pKa = 6.08GCC197 pKa = 3.36
MM1 pKa = 7.36FPLSKK6 pKa = 10.63LIISASWYY14 pKa = 8.29EE15 pKa = 3.88VHH17 pKa = 7.34GDD19 pKa = 4.04GVDD22 pKa = 3.92DD23 pKa = 5.12FNGDD27 pKa = 4.5DD28 pKa = 3.66IFALAACIEE37 pKa = 4.22GDD39 pKa = 4.59GDD41 pKa = 6.02DD42 pKa = 5.53DD43 pKa = 6.4DD44 pKa = 6.51SDD46 pKa = 4.13YY47 pKa = 11.41DD48 pKa = 3.78YY49 pKa = 11.73APAAMEE55 pKa = 5.19GDD57 pKa = 5.03DD58 pKa = 6.13DD59 pKa = 6.8DD60 pKa = 7.45DD61 pKa = 6.72DD62 pKa = 6.51DD63 pKa = 6.51GEE65 pKa = 4.41IVLPINSVRR74 pKa = 11.84LEE76 pKa = 4.11EE77 pKa = 4.11SSMWKK82 pKa = 10.27DD83 pKa = 3.07KK84 pKa = 11.14YY85 pKa = 11.05SPKK88 pKa = 10.03FDD90 pKa = 3.23GKK92 pKa = 10.18VNRR95 pKa = 11.84VDD97 pKa = 4.37RR98 pKa = 11.84KK99 pKa = 9.6NEE101 pKa = 3.8LYY103 pKa = 10.6ASRR106 pKa = 11.84VHH108 pKa = 5.38QNYY111 pKa = 9.0YY112 pKa = 10.79FMAPVFKK119 pKa = 10.81LIGLSCYY126 pKa = 9.74SNHH129 pKa = 5.34MHH131 pKa = 7.59GDD133 pKa = 3.22GDD135 pKa = 4.93DD136 pKa = 3.57IYY138 pKa = 11.54GPALNGTAMTTTFPTMTTLQAPAACTEE165 pKa = 4.16WEE167 pKa = 4.05GDD169 pKa = 4.15DD170 pKa = 3.9YY171 pKa = 12.09DD172 pKa = 4.13EE173 pKa = 4.59STTILTMILHH183 pKa = 5.29QQHH186 pKa = 7.26DD187 pKa = 4.27LMLIIVVHH195 pKa = 6.08GCC197 pKa = 3.36
Molecular weight: 21.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5E4EKL8|A0A5E4EKL8_PRUDU Protein kinase domain-containing protein OS=Prunus dulcis OX=3755 GN=ALMOND_2B006539 PE=4 SV=1
MM1 pKa = 7.54SPPPKK6 pKa = 9.88HH7 pKa = 6.6KK8 pKa = 10.29PPPPQKK14 pKa = 10.22HH15 pKa = 5.77KK16 pKa = 10.48PPPPQKK22 pKa = 10.08HH23 pKa = 5.81KK24 pKa = 10.27PPPPPTPKK32 pKa = 10.31LNPPPPPNLSPPPPPPNLRR51 pKa = 11.84PPPPNLNPPPPPKK64 pKa = 10.41SNPRR68 pKa = 11.84PPPKK72 pKa = 9.97NRR74 pKa = 11.84PPPPPQNLNPPPPPQNLNPPPPPPQKK100 pKa = 9.46RR101 pKa = 11.84TPPPPPQRR109 pKa = 11.84IPPPNLIPPPPPPQNLNPPPQKK131 pKa = 9.48RR132 pKa = 11.84TPPPPPPRR140 pKa = 11.84RR141 pKa = 11.84PPPNLIPPPPPPQNLNPPPPPPKK164 pKa = 10.16KK165 pKa = 9.5LTPPPPQPRR174 pKa = 11.84RR175 pKa = 11.84PPPNLIPPPPPRR187 pKa = 11.84RR188 pKa = 11.84PPPNLIPPPPPPQNLNPPPPPPRR211 pKa = 11.84KK212 pKa = 8.75RR213 pKa = 11.84TPPPPPARR221 pKa = 11.84RR222 pKa = 11.84PPPNLIPPPPPAPKK236 pKa = 9.93QSPGTTATTPKK247 pKa = 9.72HH248 pKa = 5.72NPPVTPHH255 pKa = 6.47LMKK258 pKa = 10.87LHH260 pKa = 6.3
MM1 pKa = 7.54SPPPKK6 pKa = 9.88HH7 pKa = 6.6KK8 pKa = 10.29PPPPQKK14 pKa = 10.22HH15 pKa = 5.77KK16 pKa = 10.48PPPPQKK22 pKa = 10.08HH23 pKa = 5.81KK24 pKa = 10.27PPPPPTPKK32 pKa = 10.31LNPPPPPNLSPPPPPPNLRR51 pKa = 11.84PPPPNLNPPPPPKK64 pKa = 10.41SNPRR68 pKa = 11.84PPPKK72 pKa = 9.97NRR74 pKa = 11.84PPPPPQNLNPPPPPQNLNPPPPPPQKK100 pKa = 9.46RR101 pKa = 11.84TPPPPPQRR109 pKa = 11.84IPPPNLIPPPPPPQNLNPPPQKK131 pKa = 9.48RR132 pKa = 11.84TPPPPPPRR140 pKa = 11.84RR141 pKa = 11.84PPPNLIPPPPPPQNLNPPPPPPKK164 pKa = 10.16KK165 pKa = 9.5LTPPPPQPRR174 pKa = 11.84RR175 pKa = 11.84PPPNLIPPPPPRR187 pKa = 11.84RR188 pKa = 11.84PPPNLIPPPPPPQNLNPPPPPPRR211 pKa = 11.84KK212 pKa = 8.75RR213 pKa = 11.84TPPPPPARR221 pKa = 11.84RR222 pKa = 11.84PPPNLIPPPPPAPKK236 pKa = 9.93QSPGTTATTPKK247 pKa = 9.72HH248 pKa = 5.72NPPVTPHH255 pKa = 6.47LMKK258 pKa = 10.87LHH260 pKa = 6.3
Molecular weight: 28.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
13217611 |
42 |
5365 |
413.9 |
46.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.686 ± 0.011 | 1.871 ± 0.007 |
5.295 ± 0.01 | 6.478 ± 0.016 |
4.184 ± 0.009 | 6.575 ± 0.013 |
2.456 ± 0.006 | 5.176 ± 0.01 |
6.15 ± 0.012 | 9.819 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.392 ± 0.005 | 4.463 ± 0.007 |
4.959 ± 0.013 | 3.795 ± 0.01 |
5.257 ± 0.012 | 8.941 ± 0.015 |
4.865 ± 0.008 | 6.554 ± 0.009 |
1.303 ± 0.005 | 2.773 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
