
Streptomyces phage Hank144
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Arquatrovirinae; Arequatrovirus; unclassified Arequatrovirus
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
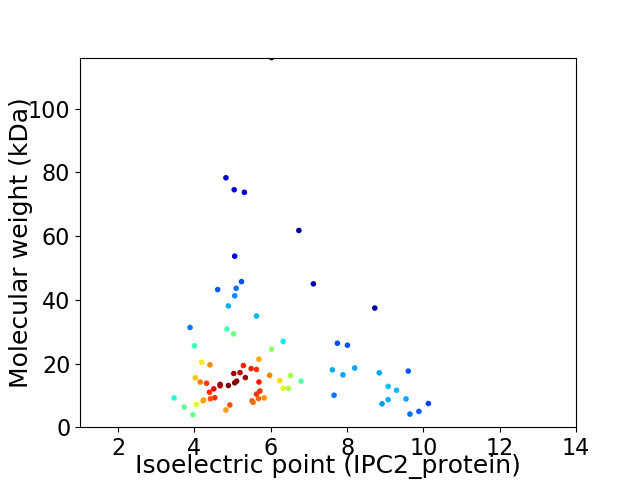
Virtual 2D-PAGE plot for 78 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
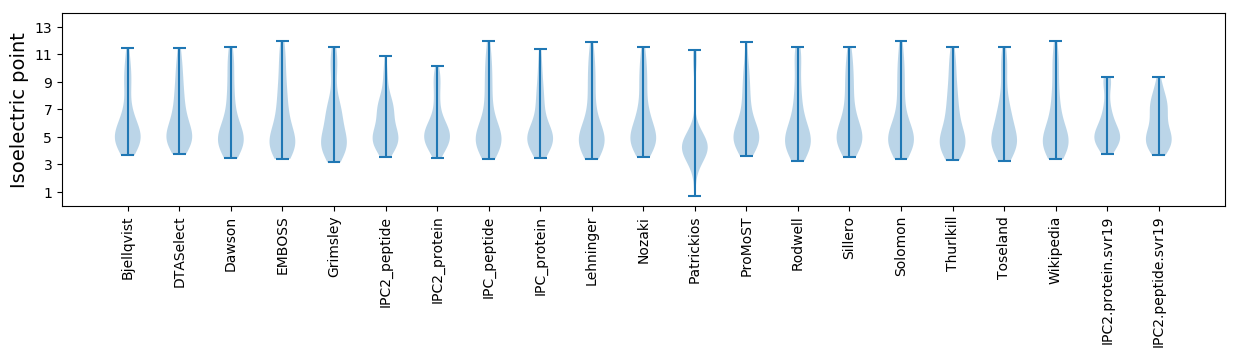
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A385DP02|A0A385DP02_9CAUD Minor tail protein OS=Streptomyces phage Hank144 OX=2301573 GN=20 PE=4 SV=1
MM1 pKa = 7.39SLDD4 pKa = 4.0FDD6 pKa = 4.29TLDD9 pKa = 4.48EE10 pKa = 4.34IAEE13 pKa = 4.19QALTRR18 pKa = 11.84PSDD21 pKa = 3.12AMFWDD26 pKa = 3.91DD27 pKa = 5.82RR28 pKa = 11.84LFTTHH33 pKa = 6.8GAILSWASRR42 pKa = 11.84GDD44 pKa = 4.19DD45 pKa = 2.94ILEE48 pKa = 4.04EE49 pKa = 4.4SNYY52 pKa = 10.35KK53 pKa = 10.23SALDD57 pKa = 4.46LIKK60 pKa = 10.76GAAGDD65 pKa = 3.84DD66 pKa = 3.87AGEE69 pKa = 4.06HH70 pKa = 6.25VIDD73 pKa = 5.18GSAGHH78 pKa = 6.62WLVGTLEE85 pKa = 4.27TIYY88 pKa = 10.65VQVYY92 pKa = 10.55ADD94 pKa = 4.0RR95 pKa = 11.84PEE97 pKa = 4.56CNTIGCEE104 pKa = 4.01EE105 pKa = 4.02EE106 pKa = 4.73AEE108 pKa = 4.12YY109 pKa = 10.47LANYY113 pKa = 7.57RR114 pKa = 11.84TGEE117 pKa = 4.03SAHH120 pKa = 6.04FCEE123 pKa = 4.44EE124 pKa = 4.04HH125 pKa = 7.13KK126 pKa = 10.9EE127 pKa = 3.97GLEE130 pKa = 3.93GSVIAEE136 pKa = 3.82ILHH139 pKa = 6.08TEE141 pKa = 4.26FEE143 pKa = 4.26PLGRR147 pKa = 11.84EE148 pKa = 3.72FTPAFVEE155 pKa = 4.13AAEE158 pKa = 4.5LLVGLQDD165 pKa = 3.62YY166 pKa = 9.33PIIDD170 pKa = 3.52EE171 pKa = 4.36SDD173 pKa = 3.18FSEE176 pKa = 4.83RR177 pKa = 11.84EE178 pKa = 3.69WKK180 pKa = 10.72AFEE183 pKa = 4.97DD184 pKa = 3.86NCSEE188 pKa = 4.12ALEE191 pKa = 4.51SASNEE196 pKa = 3.95YY197 pKa = 10.99DD198 pKa = 3.98DD199 pKa = 5.48DD200 pKa = 4.08TLEE203 pKa = 4.24EE204 pKa = 3.91ATEE207 pKa = 3.78IQNRR211 pKa = 11.84IFQDD215 pKa = 3.29SALSDD220 pKa = 3.57LFGYY224 pKa = 9.39EE225 pKa = 3.97PNAEE229 pKa = 4.21VSWEE233 pKa = 4.01RR234 pKa = 11.84VAEE237 pKa = 4.39IYY239 pKa = 10.82AEE241 pKa = 3.94YY242 pKa = 10.26RR243 pKa = 11.84DD244 pKa = 4.05AYY246 pKa = 10.5FEE248 pKa = 4.01EE249 pKa = 4.66LAYY252 pKa = 9.79EE253 pKa = 4.57AYY255 pKa = 9.58RR256 pKa = 11.84WNVLGYY262 pKa = 10.83NPDD265 pKa = 3.49QLEE268 pKa = 4.11LPIVVVIVAA277 pKa = 4.52
MM1 pKa = 7.39SLDD4 pKa = 4.0FDD6 pKa = 4.29TLDD9 pKa = 4.48EE10 pKa = 4.34IAEE13 pKa = 4.19QALTRR18 pKa = 11.84PSDD21 pKa = 3.12AMFWDD26 pKa = 3.91DD27 pKa = 5.82RR28 pKa = 11.84LFTTHH33 pKa = 6.8GAILSWASRR42 pKa = 11.84GDD44 pKa = 4.19DD45 pKa = 2.94ILEE48 pKa = 4.04EE49 pKa = 4.4SNYY52 pKa = 10.35KK53 pKa = 10.23SALDD57 pKa = 4.46LIKK60 pKa = 10.76GAAGDD65 pKa = 3.84DD66 pKa = 3.87AGEE69 pKa = 4.06HH70 pKa = 6.25VIDD73 pKa = 5.18GSAGHH78 pKa = 6.62WLVGTLEE85 pKa = 4.27TIYY88 pKa = 10.65VQVYY92 pKa = 10.55ADD94 pKa = 4.0RR95 pKa = 11.84PEE97 pKa = 4.56CNTIGCEE104 pKa = 4.01EE105 pKa = 4.02EE106 pKa = 4.73AEE108 pKa = 4.12YY109 pKa = 10.47LANYY113 pKa = 7.57RR114 pKa = 11.84TGEE117 pKa = 4.03SAHH120 pKa = 6.04FCEE123 pKa = 4.44EE124 pKa = 4.04HH125 pKa = 7.13KK126 pKa = 10.9EE127 pKa = 3.97GLEE130 pKa = 3.93GSVIAEE136 pKa = 3.82ILHH139 pKa = 6.08TEE141 pKa = 4.26FEE143 pKa = 4.26PLGRR147 pKa = 11.84EE148 pKa = 3.72FTPAFVEE155 pKa = 4.13AAEE158 pKa = 4.5LLVGLQDD165 pKa = 3.62YY166 pKa = 9.33PIIDD170 pKa = 3.52EE171 pKa = 4.36SDD173 pKa = 3.18FSEE176 pKa = 4.83RR177 pKa = 11.84EE178 pKa = 3.69WKK180 pKa = 10.72AFEE183 pKa = 4.97DD184 pKa = 3.86NCSEE188 pKa = 4.12ALEE191 pKa = 4.51SASNEE196 pKa = 3.95YY197 pKa = 10.99DD198 pKa = 3.98DD199 pKa = 5.48DD200 pKa = 4.08TLEE203 pKa = 4.24EE204 pKa = 3.91ATEE207 pKa = 3.78IQNRR211 pKa = 11.84IFQDD215 pKa = 3.29SALSDD220 pKa = 3.57LFGYY224 pKa = 9.39EE225 pKa = 3.97PNAEE229 pKa = 4.21VSWEE233 pKa = 4.01RR234 pKa = 11.84VAEE237 pKa = 4.39IYY239 pKa = 10.82AEE241 pKa = 3.94YY242 pKa = 10.26RR243 pKa = 11.84DD244 pKa = 4.05AYY246 pKa = 10.5FEE248 pKa = 4.01EE249 pKa = 4.66LAYY252 pKa = 9.79EE253 pKa = 4.57AYY255 pKa = 9.58RR256 pKa = 11.84WNVLGYY262 pKa = 10.83NPDD265 pKa = 3.49QLEE268 pKa = 4.11LPIVVVIVAA277 pKa = 4.52
Molecular weight: 31.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A385DR88|A0A385DR88_9CAUD Deoxynucleoside monophosphate OS=Streptomyces phage Hank144 OX=2301573 GN=66 PE=4 SV=1
MM1 pKa = 7.23EE2 pKa = 4.4RR3 pKa = 11.84WIPGRR8 pKa = 11.84EE9 pKa = 3.78GQYY12 pKa = 10.95AITQDD17 pKa = 3.91GVTTSYY23 pKa = 11.23VRR25 pKa = 11.84GKK27 pKa = 10.01PRR29 pKa = 11.84VLKK32 pKa = 10.2WKK34 pKa = 9.86IGTDD38 pKa = 4.18GYY40 pKa = 10.24PRR42 pKa = 11.84VAIAQKK48 pKa = 8.91WVPVHH53 pKa = 5.29TLILEE58 pKa = 4.53AWAGPRR64 pKa = 11.84PDD66 pKa = 3.38GMVVRR71 pKa = 11.84HH72 pKa = 5.93LNGDD76 pKa = 3.4PMKK79 pKa = 10.56PHH81 pKa = 7.5LSQLAWGTQSEE92 pKa = 4.28NLRR95 pKa = 11.84DD96 pKa = 3.79KK97 pKa = 10.64RR98 pKa = 11.84AHH100 pKa = 5.5GTDD103 pKa = 3.0HH104 pKa = 6.64NVNKK108 pKa = 7.47THH110 pKa = 6.63CPKK113 pKa = 10.13GHH115 pKa = 7.12EE116 pKa = 4.09YY117 pKa = 10.66TEE119 pKa = 4.68ANTLRR124 pKa = 11.84RR125 pKa = 11.84ADD127 pKa = 4.11RR128 pKa = 11.84PGHH131 pKa = 5.32RR132 pKa = 11.84ACRR135 pKa = 11.84TCNRR139 pKa = 11.84DD140 pKa = 2.65AVARR144 pKa = 11.84SRR146 pKa = 11.84QRR148 pKa = 11.84KK149 pKa = 6.95RR150 pKa = 11.84QGVAPP155 pKa = 4.45
MM1 pKa = 7.23EE2 pKa = 4.4RR3 pKa = 11.84WIPGRR8 pKa = 11.84EE9 pKa = 3.78GQYY12 pKa = 10.95AITQDD17 pKa = 3.91GVTTSYY23 pKa = 11.23VRR25 pKa = 11.84GKK27 pKa = 10.01PRR29 pKa = 11.84VLKK32 pKa = 10.2WKK34 pKa = 9.86IGTDD38 pKa = 4.18GYY40 pKa = 10.24PRR42 pKa = 11.84VAIAQKK48 pKa = 8.91WVPVHH53 pKa = 5.29TLILEE58 pKa = 4.53AWAGPRR64 pKa = 11.84PDD66 pKa = 3.38GMVVRR71 pKa = 11.84HH72 pKa = 5.93LNGDD76 pKa = 3.4PMKK79 pKa = 10.56PHH81 pKa = 7.5LSQLAWGTQSEE92 pKa = 4.28NLRR95 pKa = 11.84DD96 pKa = 3.79KK97 pKa = 10.64RR98 pKa = 11.84AHH100 pKa = 5.5GTDD103 pKa = 3.0HH104 pKa = 6.64NVNKK108 pKa = 7.47THH110 pKa = 6.63CPKK113 pKa = 10.13GHH115 pKa = 7.12EE116 pKa = 4.09YY117 pKa = 10.66TEE119 pKa = 4.68ANTLRR124 pKa = 11.84RR125 pKa = 11.84ADD127 pKa = 4.11RR128 pKa = 11.84PGHH131 pKa = 5.32RR132 pKa = 11.84ACRR135 pKa = 11.84TCNRR139 pKa = 11.84DD140 pKa = 2.65AVARR144 pKa = 11.84SRR146 pKa = 11.84QRR148 pKa = 11.84KK149 pKa = 6.95RR150 pKa = 11.84QGVAPP155 pKa = 4.45
Molecular weight: 17.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
15511 |
34 |
1100 |
198.9 |
21.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.921 ± 0.297 | 0.832 ± 0.122 |
6.55 ± 0.235 | 7.485 ± 0.427 |
2.888 ± 0.19 | 8.381 ± 0.397 |
2.018 ± 0.176 | 4.539 ± 0.32 |
4.481 ± 0.28 | 8.278 ± 0.428 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.134 ± 0.13 | 2.882 ± 0.187 |
4.861 ± 0.19 | 3.301 ± 0.149 |
6.673 ± 0.429 | 5.983 ± 0.307 |
6.273 ± 0.245 | 6.866 ± 0.199 |
1.825 ± 0.113 | 2.83 ± 0.216 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
