
Pacific flying fox faeces associated circular DNA virus-11
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.41
Get precalculated fractions of proteins
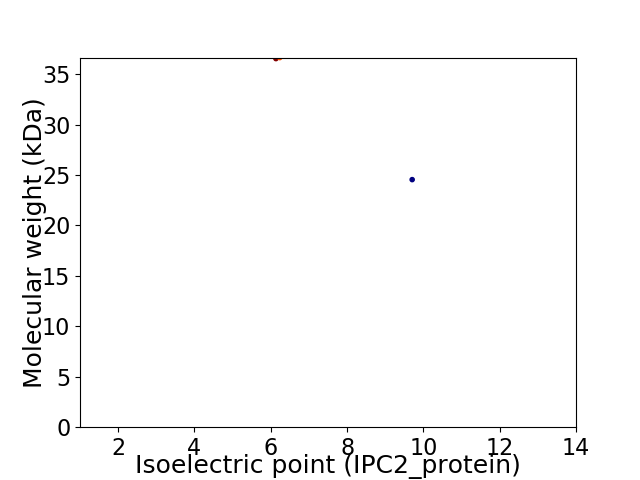
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTV1|A0A140CTV1_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-11 OX=1796006 PE=4 SV=1
MM1 pKa = 7.6SKK3 pKa = 10.4PSRR6 pKa = 11.84NFCFTLNNPDD16 pKa = 3.88DD17 pKa = 4.72EE18 pKa = 4.91YY19 pKa = 11.3TFFADD24 pKa = 4.35RR25 pKa = 11.84FPRR28 pKa = 11.84FQTQYY33 pKa = 11.43AIWQLEE39 pKa = 3.78AGEE42 pKa = 4.35NGTRR46 pKa = 11.84DD47 pKa = 3.4LQGYY51 pKa = 9.72LEE53 pKa = 4.8LSSPARR59 pKa = 11.84VSAFQSTTLKK69 pKa = 10.46GAHH72 pKa = 5.96FEE74 pKa = 4.12PRR76 pKa = 11.84KK77 pKa = 8.33GTRR80 pKa = 11.84AQARR84 pKa = 11.84DD85 pKa = 3.61YY86 pKa = 10.97CRR88 pKa = 11.84KK89 pKa = 10.0LDD91 pKa = 3.73TRR93 pKa = 11.84VDD95 pKa = 4.15GPWEE99 pKa = 4.03YY100 pKa = 10.5GTFGAGGQGRR110 pKa = 11.84RR111 pKa = 11.84SDD113 pKa = 3.93LEE115 pKa = 4.03AVEE118 pKa = 4.47QSIKK122 pKa = 10.55RR123 pKa = 11.84GASLAEE129 pKa = 3.83IAEE132 pKa = 4.18EE133 pKa = 4.19HH134 pKa = 5.18TTSFIKK140 pKa = 10.31YY141 pKa = 9.02ARR143 pKa = 11.84NISLTCEE150 pKa = 4.1ILCPTTPRR158 pKa = 11.84NPEE161 pKa = 4.09TPCEE165 pKa = 3.81VHH167 pKa = 6.93LYY169 pKa = 9.99HH170 pKa = 7.22GPSGCGKK177 pKa = 8.06TRR179 pKa = 11.84AITADD184 pKa = 3.45SLLQGSSVYY193 pKa = 10.05WKK195 pKa = 10.82DD196 pKa = 3.24NGKK199 pKa = 8.16WWDD202 pKa = 3.73GYY204 pKa = 10.13RR205 pKa = 11.84GHH207 pKa = 7.12TIVVADD213 pKa = 4.46DD214 pKa = 3.82FSGASMRR221 pKa = 11.84YY222 pKa = 8.67CDD224 pKa = 3.81FRR226 pKa = 11.84RR227 pKa = 11.84WFDD230 pKa = 3.61RR231 pKa = 11.84YY232 pKa = 8.57PCRR235 pKa = 11.84VEE237 pKa = 4.71RR238 pKa = 11.84KK239 pKa = 9.83GDD241 pKa = 3.54TTEE244 pKa = 4.34LVATRR249 pKa = 11.84FYY251 pKa = 10.73ISSTKK256 pKa = 10.65LPDD259 pKa = 3.37LWWDD263 pKa = 3.31RR264 pKa = 11.84SVVKK268 pKa = 10.05EE269 pKa = 3.98YY270 pKa = 11.06NWMEE274 pKa = 3.55IARR277 pKa = 11.84RR278 pKa = 11.84ITHH281 pKa = 6.2FHH283 pKa = 6.15TFDD286 pKa = 3.52EE287 pKa = 4.8EE288 pKa = 4.48TKK290 pKa = 10.41HH291 pKa = 5.19LHH293 pKa = 6.38SYY295 pKa = 9.46TGTDD299 pKa = 2.64QATALDD305 pKa = 4.28QYY307 pKa = 10.77CDD309 pKa = 3.65SINYY313 pKa = 7.57YY314 pKa = 10.43HH315 pKa = 6.76YY316 pKa = 11.5
MM1 pKa = 7.6SKK3 pKa = 10.4PSRR6 pKa = 11.84NFCFTLNNPDD16 pKa = 3.88DD17 pKa = 4.72EE18 pKa = 4.91YY19 pKa = 11.3TFFADD24 pKa = 4.35RR25 pKa = 11.84FPRR28 pKa = 11.84FQTQYY33 pKa = 11.43AIWQLEE39 pKa = 3.78AGEE42 pKa = 4.35NGTRR46 pKa = 11.84DD47 pKa = 3.4LQGYY51 pKa = 9.72LEE53 pKa = 4.8LSSPARR59 pKa = 11.84VSAFQSTTLKK69 pKa = 10.46GAHH72 pKa = 5.96FEE74 pKa = 4.12PRR76 pKa = 11.84KK77 pKa = 8.33GTRR80 pKa = 11.84AQARR84 pKa = 11.84DD85 pKa = 3.61YY86 pKa = 10.97CRR88 pKa = 11.84KK89 pKa = 10.0LDD91 pKa = 3.73TRR93 pKa = 11.84VDD95 pKa = 4.15GPWEE99 pKa = 4.03YY100 pKa = 10.5GTFGAGGQGRR110 pKa = 11.84RR111 pKa = 11.84SDD113 pKa = 3.93LEE115 pKa = 4.03AVEE118 pKa = 4.47QSIKK122 pKa = 10.55RR123 pKa = 11.84GASLAEE129 pKa = 3.83IAEE132 pKa = 4.18EE133 pKa = 4.19HH134 pKa = 5.18TTSFIKK140 pKa = 10.31YY141 pKa = 9.02ARR143 pKa = 11.84NISLTCEE150 pKa = 4.1ILCPTTPRR158 pKa = 11.84NPEE161 pKa = 4.09TPCEE165 pKa = 3.81VHH167 pKa = 6.93LYY169 pKa = 9.99HH170 pKa = 7.22GPSGCGKK177 pKa = 8.06TRR179 pKa = 11.84AITADD184 pKa = 3.45SLLQGSSVYY193 pKa = 10.05WKK195 pKa = 10.82DD196 pKa = 3.24NGKK199 pKa = 8.16WWDD202 pKa = 3.73GYY204 pKa = 10.13RR205 pKa = 11.84GHH207 pKa = 7.12TIVVADD213 pKa = 4.46DD214 pKa = 3.82FSGASMRR221 pKa = 11.84YY222 pKa = 8.67CDD224 pKa = 3.81FRR226 pKa = 11.84RR227 pKa = 11.84WFDD230 pKa = 3.61RR231 pKa = 11.84YY232 pKa = 8.57PCRR235 pKa = 11.84VEE237 pKa = 4.71RR238 pKa = 11.84KK239 pKa = 9.83GDD241 pKa = 3.54TTEE244 pKa = 4.34LVATRR249 pKa = 11.84FYY251 pKa = 10.73ISSTKK256 pKa = 10.65LPDD259 pKa = 3.37LWWDD263 pKa = 3.31RR264 pKa = 11.84SVVKK268 pKa = 10.05EE269 pKa = 3.98YY270 pKa = 11.06NWMEE274 pKa = 3.55IARR277 pKa = 11.84RR278 pKa = 11.84ITHH281 pKa = 6.2FHH283 pKa = 6.15TFDD286 pKa = 3.52EE287 pKa = 4.8EE288 pKa = 4.48TKK290 pKa = 10.41HH291 pKa = 5.19LHH293 pKa = 6.38SYY295 pKa = 9.46TGTDD299 pKa = 2.64QATALDD305 pKa = 4.28QYY307 pKa = 10.77CDD309 pKa = 3.65SINYY313 pKa = 7.57YY314 pKa = 10.43HH315 pKa = 6.76YY316 pKa = 11.5
Molecular weight: 36.54 kDa
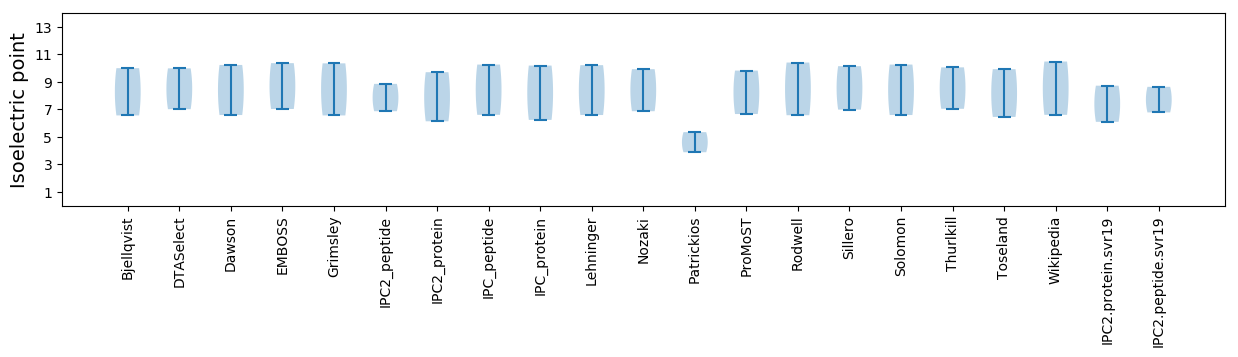
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTV1|A0A140CTV1_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-11 OX=1796006 PE=4 SV=1
MM1 pKa = 7.36RR2 pKa = 11.84RR3 pKa = 11.84NYY5 pKa = 9.85RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84SRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84SRR14 pKa = 11.84HH15 pKa = 4.28HH16 pKa = 6.16SKK18 pKa = 11.03YY19 pKa = 10.5NEE21 pKa = 4.03SQVFSITRR29 pKa = 11.84KK30 pKa = 6.99TWGTYY35 pKa = 8.28IQNTASNSTFWAGSLNASYY54 pKa = 11.38AKK56 pKa = 9.85TFTLGDD62 pKa = 3.61VPAVTEE68 pKa = 4.05FTNLFEE74 pKa = 4.72EE75 pKa = 4.42YY76 pKa = 9.43RR77 pKa = 11.84IVGIRR82 pKa = 11.84VTVNLDD88 pKa = 2.96TGINVQPTLDD98 pKa = 3.51YY99 pKa = 8.76GTWFMWVDD107 pKa = 3.48NDD109 pKa = 6.15DD110 pKa = 4.03GTQLDD115 pKa = 4.4PTTGSNFDD123 pKa = 3.97LVLQRR128 pKa = 11.84GRR130 pKa = 11.84VKK132 pKa = 10.12TFNGNDD138 pKa = 4.08GKK140 pKa = 10.92RR141 pKa = 11.84HH142 pKa = 4.66TMWVKK147 pKa = 9.89PMVSAEE153 pKa = 4.07YY154 pKa = 9.81YY155 pKa = 9.33RR156 pKa = 11.84ASGGVNAYY164 pKa = 9.37GPKK167 pKa = 9.24YY168 pKa = 10.54SPWLSTSNTGTPHH181 pKa = 6.2YY182 pKa = 10.45SFKK185 pKa = 10.34IWVRR189 pKa = 11.84NMTSNILGFNIMCKK203 pKa = 10.26YY204 pKa = 8.98YY205 pKa = 10.18LQFRR209 pKa = 11.84GVQQ212 pKa = 3.08
MM1 pKa = 7.36RR2 pKa = 11.84RR3 pKa = 11.84NYY5 pKa = 9.85RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84SRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84SRR14 pKa = 11.84HH15 pKa = 4.28HH16 pKa = 6.16SKK18 pKa = 11.03YY19 pKa = 10.5NEE21 pKa = 4.03SQVFSITRR29 pKa = 11.84KK30 pKa = 6.99TWGTYY35 pKa = 8.28IQNTASNSTFWAGSLNASYY54 pKa = 11.38AKK56 pKa = 9.85TFTLGDD62 pKa = 3.61VPAVTEE68 pKa = 4.05FTNLFEE74 pKa = 4.72EE75 pKa = 4.42YY76 pKa = 9.43RR77 pKa = 11.84IVGIRR82 pKa = 11.84VTVNLDD88 pKa = 2.96TGINVQPTLDD98 pKa = 3.51YY99 pKa = 8.76GTWFMWVDD107 pKa = 3.48NDD109 pKa = 6.15DD110 pKa = 4.03GTQLDD115 pKa = 4.4PTTGSNFDD123 pKa = 3.97LVLQRR128 pKa = 11.84GRR130 pKa = 11.84VKK132 pKa = 10.12TFNGNDD138 pKa = 4.08GKK140 pKa = 10.92RR141 pKa = 11.84HH142 pKa = 4.66TMWVKK147 pKa = 9.89PMVSAEE153 pKa = 4.07YY154 pKa = 9.81YY155 pKa = 9.33RR156 pKa = 11.84ASGGVNAYY164 pKa = 9.37GPKK167 pKa = 9.24YY168 pKa = 10.54SPWLSTSNTGTPHH181 pKa = 6.2YY182 pKa = 10.45SFKK185 pKa = 10.34IWVRR189 pKa = 11.84NMTSNILGFNIMCKK203 pKa = 10.26YY204 pKa = 8.98YY205 pKa = 10.18LQFRR209 pKa = 11.84GVQQ212 pKa = 3.08
Molecular weight: 24.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
528 |
212 |
316 |
264.0 |
30.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.682 ± 1.067 | 1.894 ± 0.796 |
5.871 ± 0.909 | 4.735 ± 1.329 |
5.114 ± 0.042 | 7.386 ± 0.354 |
2.652 ± 0.428 | 3.788 ± 0.008 |
4.167 ± 0.044 | 5.682 ± 0.276 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.705 ± 0.63 | 4.924 ± 1.731 |
3.788 ± 0.272 | 3.409 ± 0.06 |
8.523 ± 0.018 | 7.386 ± 0.354 |
9.47 ± 0.508 | 4.924 ± 1.203 |
3.03 ± 0.152 | 5.871 ± 0.146 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
