
Citrus chlorotic spot virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Dichorhavirus; Citrus chlorotic spot dichorhavirus
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
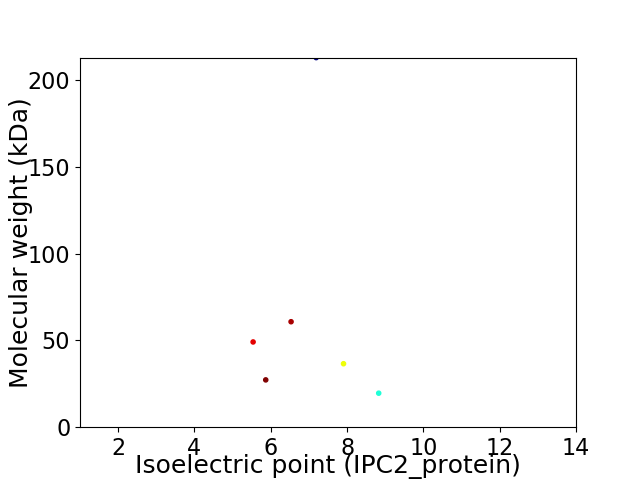
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1W6AWK5|A0A1W6AWK5_9RHAB GDP polyribonucleotidyltransferase OS=Citrus chlorotic spot virus OX=1980624 GN=ORF6 PE=4 SV=1
MM1 pKa = 7.38ARR3 pKa = 11.84YY4 pKa = 9.95ADD6 pKa = 3.42VDD8 pKa = 3.91YY9 pKa = 11.07TSVLDD14 pKa = 4.22IYY16 pKa = 11.15SDD18 pKa = 3.82TPEE21 pKa = 4.1EE22 pKa = 4.22SQVASSAPTPVTYY35 pKa = 10.59SRR37 pKa = 11.84EE38 pKa = 3.99AAVAKK43 pKa = 10.04PIVTLEE49 pKa = 4.24APPDD53 pKa = 3.47THH55 pKa = 8.51ADD57 pKa = 3.7VITLFQTIMDD67 pKa = 4.4HH68 pKa = 6.67ANSRR72 pKa = 11.84MTKK75 pKa = 10.03KK76 pKa = 10.42DD77 pKa = 3.4LLVIMSLGFCLTPPGCEE94 pKa = 4.11GPSMWSVAQGAVVQPLSMPGITAVYY119 pKa = 10.03DD120 pKa = 3.49SRR122 pKa = 11.84AAAASSTMNIDD133 pKa = 3.59PDD135 pKa = 3.83EE136 pKa = 4.93GDD138 pKa = 3.35QAEE141 pKa = 4.87DD142 pKa = 3.45SQAPPDD148 pKa = 4.59DD149 pKa = 4.12EE150 pKa = 5.79DD151 pKa = 4.41AASQCRR157 pKa = 11.84AISYY161 pKa = 9.84ICLSLMRR168 pKa = 11.84LAVKK172 pKa = 10.22RR173 pKa = 11.84VEE175 pKa = 4.28TFMKK179 pKa = 9.07GTPQLVTAYY188 pKa = 9.41HH189 pKa = 6.84ILMGDD194 pKa = 3.88HH195 pKa = 6.95SPFLASFNYY204 pKa = 9.9SRR206 pKa = 11.84GLCSNISSLFNQCDD220 pKa = 3.53DD221 pKa = 3.59MKK223 pKa = 10.53RR224 pKa = 11.84TLAHH228 pKa = 6.13HH229 pKa = 6.47CAVADD234 pKa = 4.21EE235 pKa = 4.59SHH237 pKa = 6.51HH238 pKa = 6.25NNRR241 pKa = 11.84TVHH244 pKa = 5.82GPLRR248 pKa = 11.84FLILQHH254 pKa = 6.25MDD256 pKa = 3.32LNGMVPYY263 pKa = 10.95GMYY266 pKa = 10.71VDD268 pKa = 4.28MKK270 pKa = 10.73RR271 pKa = 11.84GLPLLKK277 pKa = 10.13PGTILTWLHH286 pKa = 6.46DD287 pKa = 4.11AQVASTLMLISKK299 pKa = 9.74INKK302 pKa = 9.17DD303 pKa = 3.07HH304 pKa = 7.64DD305 pKa = 3.87RR306 pKa = 11.84TDD308 pKa = 3.09KK309 pKa = 11.19ADD311 pKa = 3.88RR312 pKa = 11.84FWRR315 pKa = 11.84YY316 pKa = 8.93CRR318 pKa = 11.84CIDD321 pKa = 3.38PGFFIEE327 pKa = 4.58LQQSRR332 pKa = 11.84CYY334 pKa = 10.61ILIARR339 pKa = 11.84MADD342 pKa = 2.74ILVRR346 pKa = 11.84GGCVNVTEE354 pKa = 4.33YY355 pKa = 11.35SDD357 pKa = 3.56PKK359 pKa = 10.14KK360 pKa = 10.94AEE362 pKa = 4.23SIKK365 pKa = 11.14DD366 pKa = 3.45KK367 pKa = 11.68ANILANAEE375 pKa = 3.95RR376 pKa = 11.84FGIEE380 pKa = 4.96FMAAYY385 pKa = 8.96NALSGTSEE393 pKa = 4.09GAGPVARR400 pKa = 11.84ALASAAVGQPTRR412 pKa = 11.84RR413 pKa = 11.84ILVPRR418 pKa = 11.84NPRR421 pKa = 11.84PQPTVNVTSNPPPQRR436 pKa = 11.84VGALDD441 pKa = 5.28SMIQDD446 pKa = 3.43QSS448 pKa = 3.21
MM1 pKa = 7.38ARR3 pKa = 11.84YY4 pKa = 9.95ADD6 pKa = 3.42VDD8 pKa = 3.91YY9 pKa = 11.07TSVLDD14 pKa = 4.22IYY16 pKa = 11.15SDD18 pKa = 3.82TPEE21 pKa = 4.1EE22 pKa = 4.22SQVASSAPTPVTYY35 pKa = 10.59SRR37 pKa = 11.84EE38 pKa = 3.99AAVAKK43 pKa = 10.04PIVTLEE49 pKa = 4.24APPDD53 pKa = 3.47THH55 pKa = 8.51ADD57 pKa = 3.7VITLFQTIMDD67 pKa = 4.4HH68 pKa = 6.67ANSRR72 pKa = 11.84MTKK75 pKa = 10.03KK76 pKa = 10.42DD77 pKa = 3.4LLVIMSLGFCLTPPGCEE94 pKa = 4.11GPSMWSVAQGAVVQPLSMPGITAVYY119 pKa = 10.03DD120 pKa = 3.49SRR122 pKa = 11.84AAAASSTMNIDD133 pKa = 3.59PDD135 pKa = 3.83EE136 pKa = 4.93GDD138 pKa = 3.35QAEE141 pKa = 4.87DD142 pKa = 3.45SQAPPDD148 pKa = 4.59DD149 pKa = 4.12EE150 pKa = 5.79DD151 pKa = 4.41AASQCRR157 pKa = 11.84AISYY161 pKa = 9.84ICLSLMRR168 pKa = 11.84LAVKK172 pKa = 10.22RR173 pKa = 11.84VEE175 pKa = 4.28TFMKK179 pKa = 9.07GTPQLVTAYY188 pKa = 9.41HH189 pKa = 6.84ILMGDD194 pKa = 3.88HH195 pKa = 6.95SPFLASFNYY204 pKa = 9.9SRR206 pKa = 11.84GLCSNISSLFNQCDD220 pKa = 3.53DD221 pKa = 3.59MKK223 pKa = 10.53RR224 pKa = 11.84TLAHH228 pKa = 6.13HH229 pKa = 6.47CAVADD234 pKa = 4.21EE235 pKa = 4.59SHH237 pKa = 6.51HH238 pKa = 6.25NNRR241 pKa = 11.84TVHH244 pKa = 5.82GPLRR248 pKa = 11.84FLILQHH254 pKa = 6.25MDD256 pKa = 3.32LNGMVPYY263 pKa = 10.95GMYY266 pKa = 10.71VDD268 pKa = 4.28MKK270 pKa = 10.73RR271 pKa = 11.84GLPLLKK277 pKa = 10.13PGTILTWLHH286 pKa = 6.46DD287 pKa = 4.11AQVASTLMLISKK299 pKa = 9.74INKK302 pKa = 9.17DD303 pKa = 3.07HH304 pKa = 7.64DD305 pKa = 3.87RR306 pKa = 11.84TDD308 pKa = 3.09KK309 pKa = 11.19ADD311 pKa = 3.88RR312 pKa = 11.84FWRR315 pKa = 11.84YY316 pKa = 8.93CRR318 pKa = 11.84CIDD321 pKa = 3.38PGFFIEE327 pKa = 4.58LQQSRR332 pKa = 11.84CYY334 pKa = 10.61ILIARR339 pKa = 11.84MADD342 pKa = 2.74ILVRR346 pKa = 11.84GGCVNVTEE354 pKa = 4.33YY355 pKa = 11.35SDD357 pKa = 3.56PKK359 pKa = 10.14KK360 pKa = 10.94AEE362 pKa = 4.23SIKK365 pKa = 11.14DD366 pKa = 3.45KK367 pKa = 11.68ANILANAEE375 pKa = 3.95RR376 pKa = 11.84FGIEE380 pKa = 4.96FMAAYY385 pKa = 8.96NALSGTSEE393 pKa = 4.09GAGPVARR400 pKa = 11.84ALASAAVGQPTRR412 pKa = 11.84RR413 pKa = 11.84ILVPRR418 pKa = 11.84NPRR421 pKa = 11.84PQPTVNVTSNPPPQRR436 pKa = 11.84VGALDD441 pKa = 5.28SMIQDD446 pKa = 3.43QSS448 pKa = 3.21
Molecular weight: 49.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6AWK1|A0A1W6AWK1_9RHAB Movement protein OS=Citrus chlorotic spot virus OX=1980624 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.14ATSSKK6 pKa = 9.51STEE9 pKa = 4.1EE10 pKa = 4.22LCTVMSVRR18 pKa = 11.84SRR20 pKa = 11.84VTLTSPSKK28 pKa = 10.66PPMDD32 pKa = 4.43AATIASMVKK41 pKa = 10.23AIGVMKK47 pKa = 10.6SKK49 pKa = 10.96ALTPEE54 pKa = 4.05GTEE57 pKa = 4.93VSGDD61 pKa = 3.2WQALITEE68 pKa = 5.0VARR71 pKa = 11.84QSKK74 pKa = 8.75PQVSGPFLTKK84 pKa = 10.48DD85 pKa = 3.47LEE87 pKa = 4.49SGNIYY92 pKa = 9.21TINMIIGGPLNVKK105 pKa = 10.0DD106 pKa = 4.13NNHH109 pKa = 5.33GRR111 pKa = 11.84VMTLRR116 pKa = 11.84SGPLYY121 pKa = 7.92DD122 pKa = 3.72TPHH125 pKa = 6.42YY126 pKa = 10.7SSNDD130 pKa = 3.04KK131 pKa = 10.73VDD133 pKa = 3.16IMLSSGKK140 pKa = 10.22ASVGISTSIATVKK153 pKa = 10.31RR154 pKa = 11.84PRR156 pKa = 11.84SSCEE160 pKa = 3.53KK161 pKa = 10.51LVNQYY166 pKa = 10.73IILPKK171 pKa = 10.57EE172 pKa = 4.51KK173 pKa = 9.88INPPAPRR180 pKa = 11.84PSGG183 pKa = 3.35
MM1 pKa = 7.14ATSSKK6 pKa = 9.51STEE9 pKa = 4.1EE10 pKa = 4.22LCTVMSVRR18 pKa = 11.84SRR20 pKa = 11.84VTLTSPSKK28 pKa = 10.66PPMDD32 pKa = 4.43AATIASMVKK41 pKa = 10.23AIGVMKK47 pKa = 10.6SKK49 pKa = 10.96ALTPEE54 pKa = 4.05GTEE57 pKa = 4.93VSGDD61 pKa = 3.2WQALITEE68 pKa = 5.0VARR71 pKa = 11.84QSKK74 pKa = 8.75PQVSGPFLTKK84 pKa = 10.48DD85 pKa = 3.47LEE87 pKa = 4.49SGNIYY92 pKa = 9.21TINMIIGGPLNVKK105 pKa = 10.0DD106 pKa = 4.13NNHH109 pKa = 5.33GRR111 pKa = 11.84VMTLRR116 pKa = 11.84SGPLYY121 pKa = 7.92DD122 pKa = 3.72TPHH125 pKa = 6.42YY126 pKa = 10.7SSNDD130 pKa = 3.04KK131 pKa = 10.73VDD133 pKa = 3.16IMLSSGKK140 pKa = 10.22ASVGISTSIATVKK153 pKa = 10.31RR154 pKa = 11.84PRR156 pKa = 11.84SSCEE160 pKa = 3.53KK161 pKa = 10.51LVNQYY166 pKa = 10.73IILPKK171 pKa = 10.57EE172 pKa = 4.51KK173 pKa = 9.88INPPAPRR180 pKa = 11.84PSGG183 pKa = 3.35
Molecular weight: 19.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3613 |
183 |
1874 |
602.2 |
67.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.757 ± 0.804 | 2.187 ± 0.184 |
6.061 ± 0.441 | 4.927 ± 0.335 |
2.851 ± 0.396 | 5.729 ± 0.405 |
3.211 ± 0.375 | 6.975 ± 0.478 |
5.84 ± 0.556 | 8.497 ± 0.488 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.318 ± 0.261 | 4.484 ± 0.284 |
4.761 ± 0.737 | 2.878 ± 0.479 |
5.369 ± 0.352 | 9.189 ± 0.542 |
5.231 ± 0.512 | 7.002 ± 0.378 |
1.605 ± 0.241 | 3.128 ± 0.377 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
