
Mycobacterium riyadhense
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycobacterium
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
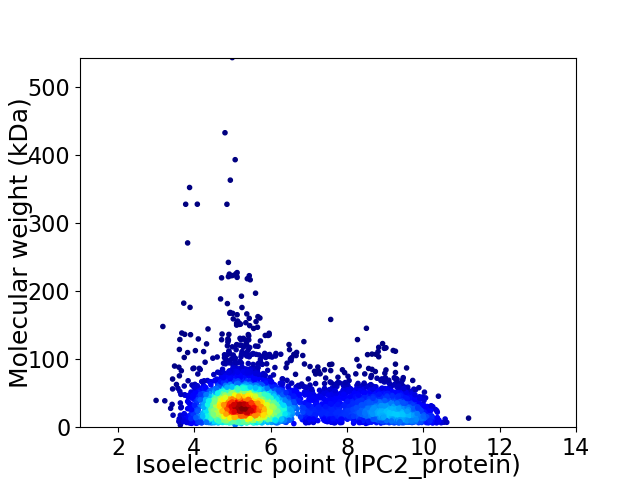
Virtual 2D-PAGE plot for 5166 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X2BE41|A0A1X2BE41_9MYCO Uncharacterized protein OS=Mycobacterium riyadhense OX=486698 GN=AWC22_04755 PE=4 SV=1
MM1 pKa = 7.52SYY3 pKa = 11.04VLAEE7 pKa = 4.48LDD9 pKa = 3.5MLAAATGDD17 pKa = 3.46MAGIGSSLSAANAAAAASTTGVLAAGGDD45 pKa = 3.92EE46 pKa = 4.31VSAAIAALFSSHH58 pKa = 5.67AQQYY62 pKa = 9.04QVLSAQATEE71 pKa = 4.45FHH73 pKa = 6.32AQFVQALNNAGGAYY87 pKa = 9.85AAAEE91 pKa = 4.19AANGAPLQTLVDD103 pKa = 5.31DD104 pKa = 4.3ILAVINAPTNVLLGRR119 pKa = 11.84PVIGDD124 pKa = 4.06GSNGAAGTGDD134 pKa = 3.19AGGAGGILWGNGGSGGSGAPGQAGGAGGSAGLLGSGGTGGAGGIGGGSGGVGGTGGWLWGNGGAGGAGGVGGLGVNGGAGGVGGSALLFGNGGFGGVGGAGAAGAVGDD242 pKa = 4.54AGTPGTTVFAGGTGGVGGDD261 pKa = 3.82GGTAGNGGAGGNGGLVFGAGGDD283 pKa = 3.74GGQGGVGGAGGTGGAGGAGGEE304 pKa = 4.18PTRR307 pKa = 11.84LGQAGGDD314 pKa = 3.82GGDD317 pKa = 3.9SGTGGNGGDD326 pKa = 3.9GGMGGAGGRR335 pKa = 11.84GSALFGTAGANGNGGAGGAGGDD357 pKa = 3.86AGAPGNGADD366 pKa = 3.82GAAGRR371 pKa = 11.84DD372 pKa = 3.38ASTPGGMGGNGGDD385 pKa = 3.57PGGVGVGGTGGAAGGVGASAGATGTDD411 pKa = 3.53GSIIAGNGGNGGHH424 pKa = 6.78GGAGFNATGSGHH436 pKa = 6.18GGSGGSGGSGGRR448 pKa = 11.84YY449 pKa = 6.5GTGGTGGAGGAAAGDD464 pKa = 4.21GNGGRR469 pKa = 11.84GGDD472 pKa = 3.56GGFGGSMGGAGGDD485 pKa = 3.71GGDD488 pKa = 3.76GGVGAGSGSGGWGGTGGHH506 pKa = 6.49HH507 pKa = 7.2LVTGTANAVGGTGGAGAAGGSTGVGGFGGVGGDD540 pKa = 3.24AKK542 pKa = 10.6IFNDD546 pKa = 3.3ASTAVATGGNGGAGGDD562 pKa = 3.98GASGGNGGDD571 pKa = 3.64GGWAYY576 pKa = 9.99TEE578 pKa = 4.92GIGNVTPGTGGHH590 pKa = 6.7GGTGTTFGGGSGGDD604 pKa = 3.17GGGAQIANGASTVVAVGGTGGAGGDD629 pKa = 4.16GIDD632 pKa = 3.28NSGFTEE638 pKa = 4.49AGHH641 pKa = 6.3GGSGGDD647 pKa = 3.73AFIDD651 pKa = 3.86NTGSTAKK658 pKa = 10.67AIGGTGGAGGSASTGTGGIGGAGGDD683 pKa = 4.2AFNSGAGSAFGGTPGAGGAGPAGGDD708 pKa = 3.48GGQGGAAYY716 pKa = 10.51ALGTGDD722 pKa = 3.53ATGHH726 pKa = 6.54AGAPGTSVGASGVGGAGGGGGDD748 pKa = 4.18ARR750 pKa = 11.84ILNSASTATATAGDD764 pKa = 4.25GGNGGDD770 pKa = 3.89GTSGGSGGYY779 pKa = 9.93GGWAYY784 pKa = 8.6TQGTGNVTAGNGGYY798 pKa = 10.4GGTGSTGGGGSGGEE812 pKa = 4.2GGNVEE817 pKa = 4.75IDD819 pKa = 3.04NDD821 pKa = 4.2AYY823 pKa = 10.93GHH825 pKa = 7.43DD826 pKa = 3.49ITGGNGGAGGTGLNQLDD843 pKa = 3.7GGFGNGGDD851 pKa = 4.16GGTATITSAGSTVNAIGGAGGAGGNATTFAASGGSGGMAINHH893 pKa = 6.24GNGNATGGFAGDD905 pKa = 4.33GGDD908 pKa = 3.9GAHH911 pKa = 6.69GGSGGSGGSAFGYY924 pKa = 10.89GNGLATGGVGGTGGNSSGVGANGGDD949 pKa = 3.71GGDD952 pKa = 3.64GGNAYY957 pKa = 10.44AHH959 pKa = 6.66VYY961 pKa = 8.19PANATEE967 pKa = 4.98GIGGAGGSGDD977 pKa = 3.8PDD979 pKa = 4.63GIAGLDD985 pKa = 4.49GITGPLL991 pKa = 3.32
MM1 pKa = 7.52SYY3 pKa = 11.04VLAEE7 pKa = 4.48LDD9 pKa = 3.5MLAAATGDD17 pKa = 3.46MAGIGSSLSAANAAAAASTTGVLAAGGDD45 pKa = 3.92EE46 pKa = 4.31VSAAIAALFSSHH58 pKa = 5.67AQQYY62 pKa = 9.04QVLSAQATEE71 pKa = 4.45FHH73 pKa = 6.32AQFVQALNNAGGAYY87 pKa = 9.85AAAEE91 pKa = 4.19AANGAPLQTLVDD103 pKa = 5.31DD104 pKa = 4.3ILAVINAPTNVLLGRR119 pKa = 11.84PVIGDD124 pKa = 4.06GSNGAAGTGDD134 pKa = 3.19AGGAGGILWGNGGSGGSGAPGQAGGAGGSAGLLGSGGTGGAGGIGGGSGGVGGTGGWLWGNGGAGGAGGVGGLGVNGGAGGVGGSALLFGNGGFGGVGGAGAAGAVGDD242 pKa = 4.54AGTPGTTVFAGGTGGVGGDD261 pKa = 3.82GGTAGNGGAGGNGGLVFGAGGDD283 pKa = 3.74GGQGGVGGAGGTGGAGGAGGEE304 pKa = 4.18PTRR307 pKa = 11.84LGQAGGDD314 pKa = 3.82GGDD317 pKa = 3.9SGTGGNGGDD326 pKa = 3.9GGMGGAGGRR335 pKa = 11.84GSALFGTAGANGNGGAGGAGGDD357 pKa = 3.86AGAPGNGADD366 pKa = 3.82GAAGRR371 pKa = 11.84DD372 pKa = 3.38ASTPGGMGGNGGDD385 pKa = 3.57PGGVGVGGTGGAAGGVGASAGATGTDD411 pKa = 3.53GSIIAGNGGNGGHH424 pKa = 6.78GGAGFNATGSGHH436 pKa = 6.18GGSGGSGGSGGRR448 pKa = 11.84YY449 pKa = 6.5GTGGTGGAGGAAAGDD464 pKa = 4.21GNGGRR469 pKa = 11.84GGDD472 pKa = 3.56GGFGGSMGGAGGDD485 pKa = 3.71GGDD488 pKa = 3.76GGVGAGSGSGGWGGTGGHH506 pKa = 6.49HH507 pKa = 7.2LVTGTANAVGGTGGAGAAGGSTGVGGFGGVGGDD540 pKa = 3.24AKK542 pKa = 10.6IFNDD546 pKa = 3.3ASTAVATGGNGGAGGDD562 pKa = 3.98GASGGNGGDD571 pKa = 3.64GGWAYY576 pKa = 9.99TEE578 pKa = 4.92GIGNVTPGTGGHH590 pKa = 6.7GGTGTTFGGGSGGDD604 pKa = 3.17GGGAQIANGASTVVAVGGTGGAGGDD629 pKa = 4.16GIDD632 pKa = 3.28NSGFTEE638 pKa = 4.49AGHH641 pKa = 6.3GGSGGDD647 pKa = 3.73AFIDD651 pKa = 3.86NTGSTAKK658 pKa = 10.67AIGGTGGAGGSASTGTGGIGGAGGDD683 pKa = 4.2AFNSGAGSAFGGTPGAGGAGPAGGDD708 pKa = 3.48GGQGGAAYY716 pKa = 10.51ALGTGDD722 pKa = 3.53ATGHH726 pKa = 6.54AGAPGTSVGASGVGGAGGGGGDD748 pKa = 4.18ARR750 pKa = 11.84ILNSASTATATAGDD764 pKa = 4.25GGNGGDD770 pKa = 3.89GTSGGSGGYY779 pKa = 9.93GGWAYY784 pKa = 8.6TQGTGNVTAGNGGYY798 pKa = 10.4GGTGSTGGGGSGGEE812 pKa = 4.2GGNVEE817 pKa = 4.75IDD819 pKa = 3.04NDD821 pKa = 4.2AYY823 pKa = 10.93GHH825 pKa = 7.43DD826 pKa = 3.49ITGGNGGAGGTGLNQLDD843 pKa = 3.7GGFGNGGDD851 pKa = 4.16GGTATITSAGSTVNAIGGAGGAGGNATTFAASGGSGGMAINHH893 pKa = 6.24GNGNATGGFAGDD905 pKa = 4.33GGDD908 pKa = 3.9GAHH911 pKa = 6.69GGSGGSGGSAFGYY924 pKa = 10.89GNGLATGGVGGTGGNSSGVGANGGDD949 pKa = 3.71GGDD952 pKa = 3.64GGNAYY957 pKa = 10.44AHH959 pKa = 6.66VYY961 pKa = 8.19PANATEE967 pKa = 4.98GIGGAGGSGDD977 pKa = 3.8PDD979 pKa = 4.63GIAGLDD985 pKa = 4.49GITGPLL991 pKa = 3.32
Molecular weight: 82.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X2AW41|A0A1X2AW41_9MYCO Dipeptidase OS=Mycobacterium riyadhense OX=486698 GN=AWC22_07430 PE=4 SV=1
MM1 pKa = 7.98IFMRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84QRR9 pKa = 11.84TLMFMMALAVIAALLMALFLGYY31 pKa = 10.48HH32 pKa = 6.44FGRR35 pKa = 11.84RR36 pKa = 11.84AGSTPAMRR44 pKa = 11.84KK45 pKa = 7.92RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84AALGGLALSLIVLMVARR67 pKa = 11.84RR68 pKa = 11.84IQQGFVVKK76 pKa = 10.42RR77 pKa = 11.84ALPGAAGGWGLKK89 pKa = 10.26LIEE92 pKa = 4.33ALQLRR97 pKa = 11.84RR98 pKa = 11.84GRR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84FHH104 pKa = 7.43RR105 pKa = 11.84PMTRR109 pKa = 11.84WAQVTRR115 pKa = 11.84AA116 pKa = 3.72
MM1 pKa = 7.98IFMRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84QRR9 pKa = 11.84TLMFMMALAVIAALLMALFLGYY31 pKa = 10.48HH32 pKa = 6.44FGRR35 pKa = 11.84RR36 pKa = 11.84AGSTPAMRR44 pKa = 11.84KK45 pKa = 7.92RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84AALGGLALSLIVLMVARR67 pKa = 11.84RR68 pKa = 11.84IQQGFVVKK76 pKa = 10.42RR77 pKa = 11.84ALPGAAGGWGLKK89 pKa = 10.26LIEE92 pKa = 4.33ALQLRR97 pKa = 11.84RR98 pKa = 11.84GRR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84FHH104 pKa = 7.43RR105 pKa = 11.84PMTRR109 pKa = 11.84WAQVTRR115 pKa = 11.84AA116 pKa = 3.72
Molecular weight: 13.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1738609 |
29 |
5181 |
336.5 |
35.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.082 ± 0.055 | 0.826 ± 0.011 |
5.846 ± 0.032 | 4.83 ± 0.037 |
3.027 ± 0.026 | 10.285 ± 0.157 |
2.186 ± 0.02 | 4.301 ± 0.026 |
2.077 ± 0.023 | 9.648 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.992 ± 0.017 | 2.688 ± 0.055 |
5.691 ± 0.032 | 3.117 ± 0.019 |
7.024 ± 0.047 | 5.549 ± 0.03 |
5.977 ± 0.028 | 8.293 ± 0.04 |
1.462 ± 0.013 | 2.102 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
